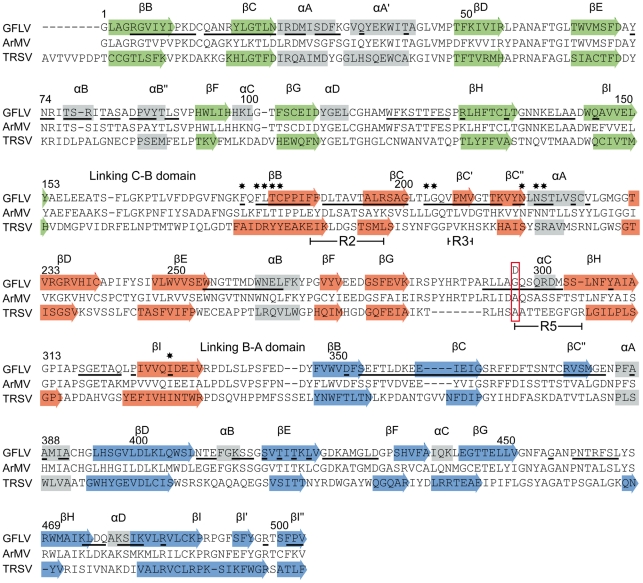
Figure 3. Alignment of the CP amino acid sequences of GFLV, ArMV and TRSV.
Secondary structures observed in GFLV and TRSV crystal structures are indicated by arrows (β strands) and grey blocks (α helices). The sequence alignment was created with Clustal X. The same color code as in Figure 2 is used for strands to indicate the three CP domains: green, red and blue for the C, B, and A domains, respectively. Residues located at the outer surface of the GFLV capsid are underlined. Residues present at position 297 in GFLV-TD, GFLV-F13, ArMV and TRSV are boxed in red. Regions R2, R3 and R5 (see [32] are indicated below the alignments. Stars indicate residues at the bottom of the putative ligand-binding pocket.