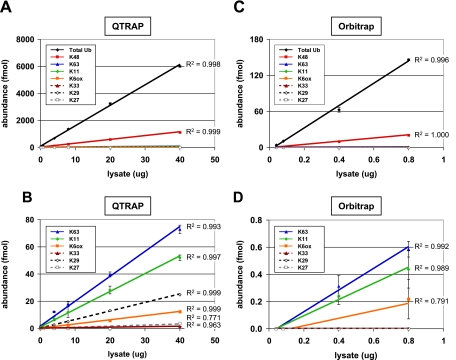
Fig. 6.
Dilution series of proteasome inhibitor-treated 293T cell lysate on multiple instrument platforms. A, a series of lysate samples ranging from 0.04 to 40 μg was prepared and analyzed on a QTRAP system in the presence of 75 fmol of Ub-AQUA peptide mixture. The amount of lysate loaded on the column is plotted against the measured abundance for total Ub and each polyUb linkage. Total Ub was calculated as the average of all loci (K6, K11, K33, K48, and K63). Error bars represent S.D. (n = 3), and R2 values are shown for each linear fit. Total Ub was detectable in all injections, increasing linearly (R2 = 0.998) from 5.7 fmol in 0.04 μg of lysate to 6004 fmol in 40 μg of lysate. Similarly, 0.9 fmol of K48 was quantified in 0.04 μg of lysate up to 1126 fmol in 40 μg of lysate. B, 100× magnification of data from A showing detection of lower abundance polyUb linkages. R2 values greater than 0.99 can be obtained from 40 μg down to a 0.4-μg injection for K63, a 0.8-μg injection for K11, and a 4.0-μg injection for K6(ox) and K29. K27 and K33 signal was only detectable from 20- or 40-μg injection levels, whereas linear polyUb chains were not detected in any of the samples. C, subset of lysate samples (with 75 fmol of Ub-AQUA peptide mixture) used in A (0.04–0.8 μg) was analyzed on the Orbitrap (n = 2). A total Ub level of 4.0 fmol was measured in 0.04 μg of lysate, increasing linearly (R2 = 0.996) up to 146 fmol in 0.8 μg of lysate. D, 225× magnification of C. K63 and K11 were detected only at 0.4- and 0.8-μg lysate eq. K6(ox) was detectable only at a 0.8-μg lysate load, accounting for an R2 value of 0.79. K33, K29, K27, and linear chains were not detectable even at the highest injection levels.