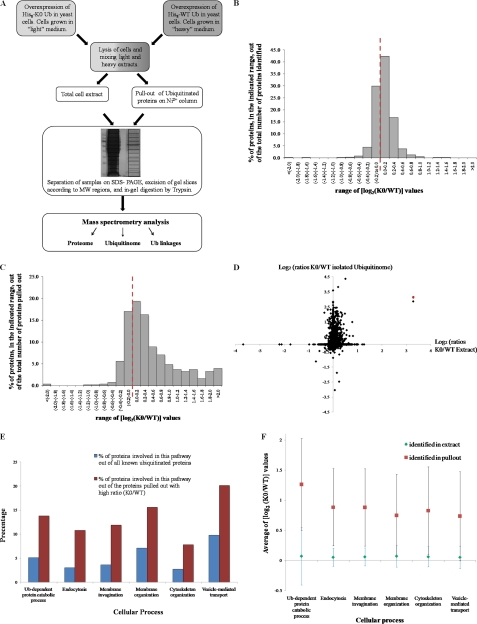
Fig. 3.
Impact of K0 Ub expression on proteome and ubiquitinome. A, Schematic description of SILAC approach used in this study. Cells expressing RGS-His8 tagged WT or RGS-His8 tagged K0 Ub were differentially isotopically labeled, mixed, lysed under denaturing conditions, and split for MW resolution of total proteome or for affinity purification of Ub-conjugates. MS/MS analysis was performed at three levels: proteins in total cell extract (proteome; panel B); proteins in pullout of ubiquitin-conjugates (ubiquitinome; panel C); and analysis of ubiquitin linkages (Fig. 4). B, 2196 different proteins were identified at high confidence in whole cell extracts of K0 Ub and WT Ub expressing cells. Enrichment factors were calculated from intensity of MS signals (log2(K0/WT); supplemental Table S2). Bars represent frequency of proteins (%) in each enrichment factor range. The red dotted line represents a ratio of unity (log21 = 0); i.e. proteins whose ratio was unaltered by expression of K0 Ub. C, A total of 856 different proteins were identified by Ub-affinity pullout (supplemental Table S1). Enrichment factors were calculated and displayed as in panel B. D, The enrichment factor for each protein in the Ub pullout (y axis) was plotted against its enrichment factor in whole cell extract (x axis). The two dimensional plot indicates the K0 Ub-induced perturbation on ubiquitination status of each substrate relative to changes in its cellular abundance. A red asterisk represents the K0 Ub protein, which was added to the database of all possible yeast translated open reading frames. E, Biological pathways significantly enriched with ubiquitinated proteins upon K0 Ub expression. Identified proteins were classified into biological pathways using the AmiGO program. Ubiquitinated proteins with a greater than twofold increase in K0 Ub expressing cells were categorized according to biological pathways. The relative portion that each category makes up out of total ubiquitinated proteins is shown in red bars. Blue bars represent the portion that these pathways make up of known ubiquitinated targets (taken from supplemental Table S3). F, To emphasize the enrichment of ubiquitinated proteins in specific biological categories the span of enrichment factors for all proteins identified in the Ub pullout is displayed for each category (surrounding the average enrichment ratio marked as red squares). For comparison, the enrichment factors of these same proteins in whole cell extract are also shown along with the average enrichment in extract of proteins belonging to each category (green diamonds). The comparison highlights that proteins belonging to these categories accumulate as ubiquitinated conjugates in K0 Ub expressing cells.