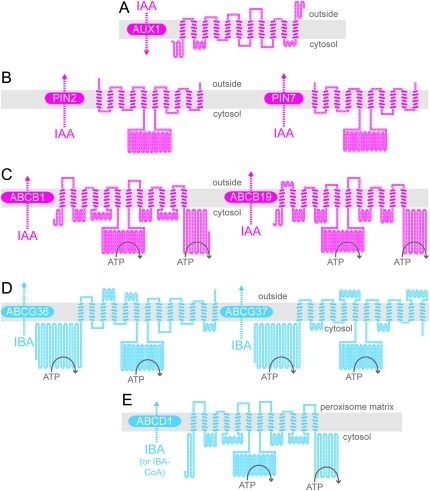
Figure 2.
Predicted Topologies of IAA Carrier Proteins that Do Not Transport IBA (A–C) and Proteins Demonstrated (D) or Suggested (E) to Transport IBA.
Schematic diagrams illustrating the predicted topologies of AUX1 (A), PIN2 and PIN7 (B), ABCB1 and ABCB19 (C), ABCG36 and ABCG37 (D), and ABCD1 (E) based on the outputs of ARAMEMNON (http://aramemnon.botanik.uni-koeln.de; Schwacke et al., 2003) and TOPO2 (www.sacs.ucsf.edu/TOPO2). Each amino acid residue is represented by a circle; filled circles represent residues predicted to span the membrane (gray rectangle). Positions of nucleotide-binding domains in the ABC proteins are schematized by ATP hydrolysis. For AUX1, the ARAMEMNON TmConsens prediction of transmembrane domains predicted 10 transmembrane domains with strong scores and one additional transmembrane domain with a weak score. Because AUX1 was experimentally shown to have 11 transmembrane domains (Swarup et al., 2004), all 11 transmembrane domains are depicted in the diagram. We used the ARAMEMNON TmConsens predictions for PIN2, PIN7, ABCB1, ABCB19, and ABCG37 to create the corresponding models, and the ARAMEMNON TmHMM_v2 prediction (Sonnhammer et al., 1998) to create the ABCG36 diagram. The ARAMEMNON MemSat_v3 (Jones et al., 1994) predicted 13 transmembrane domains for ABCD1. However, because the ATPase domains of ABCD1 are cytosolic (Nyathi et al., 2010), we did not include the first predicted transmembrane domain in the ABCD1 diagram.