Abstract
We investigated the mechanisms of uptake of 2-chlorobenzoate (2-CBa) and 2-hydroxybenzoate (2-HBa) by Pseudomonas huttiensis strain D1. Uptake was monitored by assaying intracellular accumulation of 2-[UL-ring-14C]CBa and 2-[UL-ring-14C]HBa. Uptake of 2-CBa showed substrate saturation kinetics with an apparent Km of 12.7 ± 2.6 μM and a maximum velocity (Vmax) of 9.76 ± 0.78 nmol min−1 mg of protein−1. Enhanced rates of uptake were induced by growth on 2-CBa and 2-HBa, but not by growth on benzoate or 2,5-di-CBa. Intracellular accumulations of 2-CBa and 2-HBa were 109- and 42-fold greater, respectively, than the extracellular concentrations of these substrates and were indicative of uptake mediated by a transporter rather than driven by substrate catabolism (“metabolic drag”). Results of competitor screening tests indicated that the substrate range of the transporter did not include other o-halobenzoates that serve as growth substrates for strain D1 and for which the metabolism was initiated by the same dioxygenase as 2-CBa and 2-HBa. This suggested that multiple mechanisms for substrate uptake were coupled to the same catabolic enzyme. The preponderance of evidence from tests with metabolic inhibitors and artificial electrochemical gradients suggested that 2-CBa uptake was driven by ATP hydrolysis. If so, the 2-CBa transporter would be the first of the ATP binding cassette type implicated in uptake of haloaromatic acids.
Biodegradation of aromatic acids is a process important to both cycling of natural organic matter and dissipation of xenobiotic compounds. The latter of these includes compounds introduced directly into the environment (e.g., as pesticides) as well as those formed from the biodegradation of other compounds (e.g., transformation of polychlorinated biphenyls to chlorobenzoates). The metabolism of aromatic acids has thus been the subject of much research, the majority of which has focused on activities of catabolic enzymes. However, the uptake mechanisms by which the substrate is delivered to these intracellular enzymes have received comparatively little study, especially those that may be operative with chlorinated compounds.
Uptake of aromatic acids by bacteria can be transporter mediated or driven by diffusion across intracellular/extracellular gradients of pH and substrate concentration (20). Establishment and maintenance of concentration gradients requires the intracellular substrate concentration to be kept low relative to that of the external environment, which may be achieved by rapid transformation of the imported compound to metabolic intermediates (11, 26, 37). Thus, in this case uptake is effectively driven by the activity of catabolic enzymes, and this “metabolic drag” (37) mechanism has been proposed for the uptake of benzoate (11) and 4-hydroxybenzoate (4-HBa) (26) in Rhodopseudomonas palustris and uptake of 4-HBa by Rhizobium leguminosarum (37).
Transporter-mediated uptake has been demonstrated for several aromatic acids with a variety of bacteria. These compounds and organisms include benzoate (Pseudomonas putida [35], Acinetobacter sp. ADP1 [9]), 4-HBa (Klebsiella pneumoniae [5], Acinetobacter sp. [2]), Klebsiella planticola [3], P. putida [12]), protocatechuate (P. putida [28]), mandelate (P. putida [17]), phenylacetate (P. putida [33]), 4-hydroxyphenylacetate (Escherichia coli [31], K. pneumoniae [4]), and phthalate and 4-methylphthalate (Burkholderia fungorum [8, 32]). Chlorinated aromatic compounds for which transporters have been demonstrated are 4-chlorobenzoate (coryneform bacterium strain NTB-1 [10]), 2,4-dichlorophenoxyacetate (Ralstonia eutropha [24], Sphingomonas herbicidovorans [38]), and dichlorprop (S. herbicidovorans [38]). For most of these, the carrier that mediated uptake was demonstrated or implicated as being a secondary transporter, which utilized energy stored in electrochemical gradients of the cytoplasmic membrane to drive substrate movement. For only two of the compounds and organisms described above was uptake proposed to be mediated by an ABC-type primary transporter (energized by ATP hydrolysis): 4-hydroxyphenylacetate in K. pneumoniae strain M5a1 (4) and 4-HBa in Acinetobacter sp. strain BEM2 (2).
Pseudomonas huttiensis strain D1 acquired the ability to utilize a variety of ortho-halobenzoates by horizontal transfer of genes from Pseudomonas aeruginosa strain JB2 (29). Biodegradation genes acquired by strain D1 from strain JB2 that have been characterized to date include those encoding a salicylate 5-hydroxlase (hybABCD [15]) and a 2-halobenzoate 1,2-dioxygenase (ohbAB [14]). Also identified previously was a cluster of genes (hybEFG) immediately downstream of hybABCD whose products had significant identities to components of an ABC-type transporter (15). The potential activity of this putative transporter in mediating uptake of salicylate or 2-chlorobenzoate (2-CBa) was not determined.
In the present study, we focused on P. huttiensis strain D1 and investigated the uptake mechanisms for 2-CBa and 2-hydroxybenzoate (2-HBa). Our objectives were to determine if uptake was an active transporter-mediated process, define the kinetic parameters of transport, elucidate the substrate range of the transport system, link transport energetics to either ATP hydrolysis or electrochemical gradients, and determine if the putative ABC transporter encoded by hybEFG was involved with 2-CBa or 2-HBa uptake.
MATERIALS AND METHODS
Cultures, culture conditions, and DNA manipulations.
The origin and characteristics of P. huttiensis strain D1 are described elsewhere (14, 15, 29). Strain D1 was grown in batch or continuous culture on mineral salts medium, pH 7.0, supplemented with 500 mg of 2-CBa, 2-HBa, or other carbon substrates liter−1 as described elsewhere (13). The majority of experiments were done with continuous-culture cells grown on 2-CBa at a dilution rate of 0.086 h−1. The μmax was 0.158 ± 0.037 h−1 as determined from duplicate washout experiments. Batch-culture cells were used for experiments on induction of the transporter by growth on substrates other than 2-CBa and were harvested in late log phase. Isolation of genomic DNA, restriction enzyme digestions, probe preparation, and Southern hybridization procedures were done as described previously (15).
Uptake assays.
Radiolabeled compounds used in these assays were 2-[U-ring-14C]CBa (purity, 96%; specific activity, 29 mCi mmol−1) obtained from Sigma Chemical Co. (St. Louis, Mo.) and 2-[U-ring-14C]Ba (purity, 99%; specific activity, 15 mCi mmol−1) from American Radiolabeled Chemicals, Inc. (St. Louis, Mo.). For 2-CBa uptake assays, a hot-cold stock solution (specific activity, 2.9 mCi mmol−1) was prepared in sterile 50 mM NaKPO4 buffer with 100 μM 2-[14C]CBa and 900 μM unlabeled 2-CBa (purity, 98%; Aldrich, Milwaukee, Wis.). A hot-cold stock solution of 2-HBa (specific activity, 3.0 mCi mmol−1) contained 200 μM 2-[14C]HBa with 800 μM unlabeled 2-HBa (purity, 99%; Sigma).
Cells were harvested by centrifugation (6,000 × g, 10 min), washed once with assay buffer (50 mM NaKPO4 [pH 7.0]), and resuspended in this buffer to an optical density corresponding to 0.05 ± 0.01 mg of total cellular protein ml−1. For the assays, cell suspension (1 ml) was incubated at 30°C for 10 min on a shaker rotating at 120 rpm, and reactions were initiated by addition of the hot-cold stock solutions. For kinetic determinations, the concentration of 2-[14C]CBa was varied from 2.5 to 60 μM, all other reactions contained 10 μM (29 nCi) of 2-[14C]CBa or 2-[14C]HBa (30 nCi). Aliquots (200 μl) were removed from the reaction mixture at 30-s intervals for 2.5 min. Cells were filtered onto nitrocellulose membranes (0.45-μm-pore-size diameter; Whatman International Ltd., Maidstone, England) on a vacuum manifold and rinsed with 4 ml of assay buffer. Reactions were quenched by placing the filters in ScintiSafe scintillation cocktail preheated to 70°C. Membranes were removed from the manifold at 30-s intervals, such that each remained on the manifold for 2.5 min after cells were filtered onto it. The delay resulted in 24% ± 6% loss of 14C activity on the membranes, presumably reflecting mineralization of 2-[14C]CBa and 2-[14C]HBa.
Radioactivity on the filters was counted on a RackBeta liquid scintillation counter (LKB Wallac, Turku, Finland). Uptake rates for each reaction were calculated from the slope of the linear regression of total 14C activity in the cells versus time. For all assays, the linear regression extrapolated to time zero did not pass through the origin. Due to limitations of the sampling methods, however, measurements before 0.5 min were not possible. Thus, unless otherwise indicated, the slope of the line between 0.5 and 2.0 or 2.5 min was used to calculate uptake rates. All transport rates and ATP levels were normalized to total protein present in the reaction, which was determined by a modified Bradford method (7). Statistical differences between uptake rates were assessed by Student's t test.
Values for Vmax and Km were estimated by nonlinear regression fitting to the Michaelis-Menten equation. This was done using the Solver function of Microsoft Excel to minimize the sum of the squared error between measured and calculated uptake rates for each 2-CBa concentration. Goodness-of-fit was evaluated by the coefficient of determination.
Extraction and analysis of intracellular substrate pools.
Uptake assay mixtures were prepared as described above, but the entire 1-ml volume was sampled 1 min after the addition of 2-[14C]CBa or 2-[14C]HBa. The assay was repeated eight times, and the filters were pooled. Analysis was based on a procedure described by Miguez et al. (27). The filters were immediately placed in a vial containing 9 ml of hot water (preheated to 90°C). Filtrate from each of the reactions was collected in a scintillation cocktail in order to determine the extracellular concentration of substrate. Filters were incubated at 90°C for 15 min with occasional vortex mixing, and then the water was decanted into a clean vial. A second 9-ml portion of hot water was added to the filters, the vials were incubated at 90°C for 15 min, and this extract was combined with the first. Approximately 4% of the total 14C added remained on the filters after the two hot water extractions. The pooled, hot water extracts from eight assays were acidified with 5 N H2SO4 to pH 2 and extracted twice with an equal volume of ethyl acetate. The organic phase was collected and evaporated to ca. 200 μl with a stream of nitrogen, and the final volume was measured. Aliquots from the aqueous and organic fractions were analyzed by liquid scintillation counting.
Thin-layer chromatography was used to quantify amounts of 2-[14C]CBa and 2-[14C]HBa extracted from the cells. Aliquots (10 μl) of extract were spotted onto a silica gel plate (type 60A; Whatman International Ltd.) along with 50 nmol of unlabeled catechol, 2-CBa, or 2-HBa as a standard, which was included to visualize spot migration. Plates were developed with a hexane-ethyl acetate-acetic acid (90:5:5 [vol/vol/vol]) solvent system. The 2-CBa, 2-HBa and catechol spots were visualized under UV light, excised from the plate, and placed in a scintillation vial for measurement of radioactivity.
For determination of cell dry weight, four aliquots (3 ml each) of cell suspension were filtered through oven-dried Metricel filters (0.45-μm-pore-size diameter; Pall Gelman Laboratory, Ann Arbor, Mich.) and rinsed with 5 ml of double-distilled water. The filters were dried at 105°C for 2 days and cooled in a desiccator, and cell dry weight was calculated from the difference between the final and initial weights (0.83 ± 0.13 mg). Intracellular volume was estimated by assuming 1.5 μl of fluid volume mg−1 of cell dry weight (25, 27, 38), giving a total intracellular fluid volume of cells used in the assay of 1.25 ± 0.19 μl.
Competitor analyses and metabolic inhibitor tests.
Selected substituted benzoates were examined as potential competitors of 2-CBa uptake. Test competitors were added to a final concentration of 10 μM at the initiation of the uptake assay. For metabolic inhibitor tests, cells were treated with EDTA to increase their permeability to inhibitors (19). To do so, cells were washed once with 100 mM Tris (pH 7.0) and then resuspended in this buffer. After gentle shaking for 2 min at 30°C, EDTA was added to a 1 mM concentration and the cells were incubated for an additional 5 min at 30°C. Cells were diluted with 40 ml of inhibitor assay buffer (described below) and centrifuged immediately at 6,000 × g for 10 min. Treatment with EDTA had no significant effect (P < 0.01) on 2-CBa uptake rates. Inhibitors tested were arsenate, vanadate, N,N′-dicyclohexylcarbodiimide (DCCD) (Sigma), KCN, 2,4-dinitrophenol (Sigma), valinomycin (Sigma), nigericin (Sigma), and carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma). The phosphate analogs (potential inhibitors of ATPase) arsenate and vanadate required a phosphate-free assay mixture, and so solutions were buffered with 10 mM 2-(4-morpholino)-ethane sulfonic acid (pH 7.0). For consistency, all other inhibitors were also tested using 2-(4-morpholino)-ethane sulfonic acid buffer, except for tests with valinomycin and nigericin. These compounds require K+ for activity and were examined in the standard assay buffer. Solutions of DCCD, CCCP, valinomycin, and nigericin were prepared in ethanol; addition of 10 μl of ethanol alone to the assay mixture had no significant effect (P < 0.01) on uptake rates.
Extraction of ATP from cell suspensions was done by using the perchloric acid method (19). Ice-cold 24% (vol/vol) perchloric acid (250 μl) was added to 500 μl of cell suspension, which contained the same density of cells used in the transport assay. The mixtures were incubated on ice for 20 min and centrifuged (16,000 × g, 2 min), and the supernatants (500 μl) were neutralized with 125 μl of 4 M KOH and 125 μl of 2 M KHCO3. The mixture was incubated on ice for 30 min and centrifuged as described above. Supernatants were stored at −20°C for ≤ 48 h until analyzed for ATP. To quantify ATP, 100 μl of Enliten luciferase-luciferin reagent (Promega, Madison, Wis.) was added to 50 μl of extract and 100 μl of 10 mM Tris (pH 8.0). Resultant light emissions were quantified with a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, Calif.), and relative light units were converted to ATP concentration by reference to an ATP standard curve.
Artificially induced electrochemical gradients.
The methods used to dissipate electrochemical gradient (Δp) and to artificially induce this or its components (ΔpH or ΔΨ, for electrical potential gradient) were based on those described by Groenewegen et al. (10). Continuous-culture cells were washed once in assay buffer and then resuspended in K-acetate infusion solution (50 mM K2PO4 [pH 7.0], 100 mM K-acetate). Valinomycin was added to a 5 μM concentration, and cells were incubated for 1 h at 4°C to allow infusion of K+ and acetate into the cells and thereby dissipate Δp. The cells were washed once with, and resuspended in, the infusion solution. Next, 20 μl of the cell suspension was added to 1 ml of either of the following: (i) infusion solution (control, no Δp), (ii) 50 mM Na2PO4 (pH 7.0)-100 mM Na-HEPES (cytoplasm becomes negatively charged and alkaline, reestablishes Δp), (iii) 50 mM K2PO4 (pH 7.0)-100 mM K-HEPES (cytoplasm becomes alkaline, reestablishes ΔpH), (iv) 50 mM Na2PO4 (pH 7.0)-100 mM Na acetate (cytoplasm becomes negatively charged, reestablishes ΔΨ). Gradient inducement was applied to cells incubated under aerobic and anaerobic conditions. For the latter, aliquots of the four solutions described above were boiled, sealed immediately in 20-ml serum vials, and cooled. These solutions were then flushed with oxygen-free nitrogen gas for 15 min. The solutions and cells were anaerobically transferred with gas-tight syringes to a sealed reaction vial and then flushed with nitrogen for 10 min. Uptake assays were done as described above with aliquots that were drawn from the mixtures with a gas-tight syringe.
RESULTS AND DISCUSSION
Uptake of 2-CBa and 2-HBa: rates, kinetics, and induction.
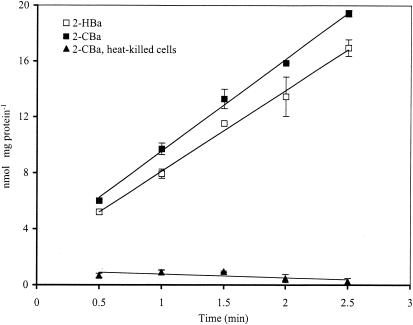
For strain D1, accumulation of 2-[14C]CBa and 2-[14C]HBa by 2-CBa-grown, continuous-culture cells was linear with respect to time for at least 2 min (Fig. 1). Average uptake rates were 4.9 ± 1.4 nmol of 2-[14C]CBa min−1 mg of protein−1 (mean of 27 assays) and 6.16 ± 0.58 nmol of 2-[14C]HBa min−1 mg of protein−1 (mean of four assays). Heat-killed (80°C, 10 min) cells retained a relatively small amount of 2-[14C]CBa (0.6 nmol mg of protein−1), which was presumed to reflect sorption and passive diffusion into the cells. The parent strain of D1, strain D, had no detectable uptake of 2-CBa (data not shown), suggesting that uptake activity was acquired from strain JB2 with the horizontal gene transfer event. Uptake of 2-CBa was observed in strain JB2 (data not shown). But the uptake was more pronounced for strain D1; all further physiological characterization of uptake was thus done with strain D1.
FIG. 1.
Uptake of 2-CBa and 2-HBa by live and heat-killed cells of P. huttiensis strain D1. Cells used in the experiment were harvested from a continuous culture growing on 2-CBa. Each data point is the mean value from three replicate assays, and error bars indicate the standard deviation of the means.
Uptake of 2-CBa followed saturation kinetics (data not shown), indicating that the process was not driven by diffusion. Nonlinear fit of data to the Michaelis-Menten equation (coefficient of determination = 0.80) gave a Vmax of 9.76 ± 0.78 nmol min−1 mg of protein−1 and apparent transport affinity (Km) of 12.7 ± 2.6 μM. The Km and Vmax of 2-CBa transport were similar to those reported for other ATP-dependent and secondary transport systems that mediate uptake of aromatic acids (2-5, 10, 24, 25, 27, 28, 31, 38). While exhibition of saturation kinetics can be used to rule out diffusion alone as the force driving uptake, the data do not distinguish between metabolic drag-driven versus transporter-mediated uptake processes.
Uptake of 2-CBa was enhanced by growth on 2-CBa or 2-HBa, indicating that the system driving uptake (catabolic enzymes or transporter) was inducible. These tests were done with cells from batch cultures, which, when grown on 2-CBa, accumulated 2-[14C]CBa at 1.56 ± 0.11 nmol min−1 mg of protein−1. In batch culture, there was no significant effect of growth phase on 2-CBa uptake rates (data not shown). There was, however, significant variability between batches in transporter activity. We also found that 2-CBa uptake rates of batch-culture-grown cells were always less than those of cells from continuous culture. Differences between batch- and continuous-culture cells in 2,5-DiCBa degradation kinetics were observed by van der Woude and colleagues (36); the physiological basis for the differences observed in the present study and that reported by those investigators remains unknown. Uptake rates of 2-CBa induced by growth on 2-HBa (1.36 ± 0.10 nmol min−1 mg of protein−1) were not significantly different (P < 0.01) from those induced in 2-CBa-grown cells. In contrast, rates of 2-CBa uptake were significantly lower (P < 0.01) for cells grown on glycerol (uptake rate in nmol min−1 mg of protein−1, 0.50 ± 0.11), 2,5-diCBa (0.46 ± 0.13), and benzoate (0.58 ± 0.24). Low levels of uptake in cells grown on these substrates could have reflected diffusion and/or basal-level transporter expression and activity. They were likely not attributable to induction and de novo transporter synthesis during the assay, since adding 50 μg of chloramphenicol ml−1 to the assay mixture did not affect 2-[14C]CBa uptake rates (data not shown). The lack of transporter induction by 2,5-diCBa was noteworthy, since catabolism by strain D1 of 2,5-diCBa and 2-CBa was presumably initiated by the same o-halobenzoate-1,2-dioxygenase (14). This finding, along with those from competitor screening tests reported below, indicated that the substrate spectrum of the transporter mediating 2-CBa uptake differed from that of the dioxygenase-initiating metabolism of this compound.
Analysis of intracellular/extracellular concentration gradients.
For the 2-CBa experiment, a total of 105 nCi (36 nmol of 2-CBa; extracellular concentration, 4.6 μM) was captured in the filtrate, and 60 nCi of 14C-labeled compound was extracted from the cells by hot water. The overall recovery of radioactivity was 71.1%; mineralization presumably accounted for the majority of the balance. Most of the 14C activity in the hot water extracts (72.5% ± 1.1%) remained in the aqueous phase after acidification and extraction with ethyl acetate. The amount of 2-CBa in the ethyl acetate extract (representing the intracellular pool) was 0.62 ± 0.29 nmol (3.0% ± 1.1% of the hot water-extracted 14C), giving a calculated intracellular concentration of 2-CBa of 500 μM. The intracellular pool of catechol, the first product of 2-CBa catabolism, was 0.18 ± 0.02 nmol (0.9% of the hot water-extracted 14C). The majority of the 14C activity in the ethyl acetate (ca. 10% of the hot water-extracted 14C) remained at the spot origin and was not identified.
In the 2-HBa experiment, 150 nCi (50 nmol of 2-HBa; extracellular concentration, 6.2 μM) was captured in the filtrate, and 50 nCi of 14C activity was extracted from cells by hot water (overall recovery, 83.3%). Partitioning of 14C in the hot water extract was similar to that observed in the 2-CBa experiment, with most of the radioactivity (66.7% ± 0.2%) remaining in the aqueous phase after ethyl acetate extraction. The amounts of 2-HBa and catechol extracted from the cells were 0.33 ± 0.07 nmol (1.9% of the hot water-extracted 14C) and 0.12 ± 0.03 nmol (1.7% of the hot water-extracted 14C), respectively. The intracellular concentration of 2-HBa was 260 μM.
The intracellular/extracellular concentration ratio) for 2-CBa was 109, while that for 2-HBa was 42, indicating that both substrates were actively accumulated against their concentration gradients. Uptake was thus not driven by metabolic drag but was instead mediated by a transport system. In other bacteria, intracellular accumulations of aromatic acid growth substrates have been previously reported, with the intracellular/extracellular concentration ratio ranging from 8 to 134 (27, 35, 38). In the present study, determinations of the intracellular pools were made on cells continuously growing on 2-CBa at ca. 54% of their maximum rate, and the activity of haloaromatic acid degraders may vary as a function of growth rate as well as other environmental parameters, such as the dissolved oxygen levels (21-23, 34, 36). A next step in this research would be to determine for strain D1 how transporter and catabolic enzyme activity vary as a function of these parameters and the consequent effect on intracellular substrate pools.
Competitors of 2-CBa uptake.
A significant reduction (P < 0.01) of 2-[14C]CBa uptake rates was noted with the additions of several ortho- and meta-substituted benzoates (Table 1). Uptake was reduced most dramatically by 3-chlorobenzoate (3-CBa) and 2-fluorobenzoate (2-FBa). In the halogen series, the relative level of inhibition increased with increasing electronegativity of the substituent. 2-Methylbenzoate, 2-hydoxybenzoate, and 3-bromobenzoate also significantly decreased 2-CBa uptake rates, but to a lesser extent than did the other compounds. 2-Nitrobenzoate, benzoate, 2-iodobenzoate, para-substituted benzoates, and di- and tri-CBa had no significant effect (P < 0.01) on 2-CBa uptake rates.
TABLE 1.
Examination of benzoates as competitors of 2-CBa uptake by 2-CBa-grown cells of strain D1
| Test competitora | Metabolism of competitorb | Uptake rate (%)c |
|---|---|---|
| None | NA | 100 |
| Benzoate | + | 92.4 ± 11.6 |
| Substituted benzoates | ||
| ortho | ||
| Fluoro- | + | 30.1 ± 0.2* |
| Chloro- | + | 51.8 ± 3.3* |
| Bromo- | + | 83.3 ± 6.3 |
| Iodo- | + | 99.2 ± 4.4 |
| Hydroxy- | + | 69.8 ± 3.2* |
| Methyl- | + | 63.4 ± 12.4* |
| Nitro- | ND | 107.7 ± 2.7 |
| meta | ||
| Chloro- | − | 29.0 ± 2.0* |
| Bromo- | ND | 53.3 ± 4.4* |
| Hydroxy- | ND | 108.2 ± 4.4 |
| para | ||
| Chloro- | − | 110.1 ± 6.9 |
| Bromo- | ND | 122.0 ± 15.1 |
| Hydroxy- | ND | 115.9 ± 16.7 |
| ortho and meta | ||
| 2,3-Dichloro- | + | 89.1 ± 14.8 |
| 2,5-Dichloro- | + | 107.0 ± 17.3 |
| 2,3,5-Trichloro- | + | 104.6 ± 2.0 |
Competitor and 2-[14C]CBa added in equal molar amounts (10 μM).
Indication of whether the competitor molecule was previously determined (13, 14, 29) to be metabolized by 2-CBa-grown cells: +, competitor metabolized; −, competitor not metabolized; NA, not applicable; ND, not determined in previous experiments.
100% = 6.39 ± 1.39 nmol min−1 mg of protein−1. Each value is the mean of two replicate assays ± standard deviation of the mean. *, values significantly different (P < 0.01) from those of the control lacking a competitor.
We regarded compounds that inhibited uptake of 2-CBa as being recognized by the carrier; recognition may or may not include translocation into the cell. Notably, by this criterion the affinity of the transporter for 3-CBa and 2-FBa was greater than that for 2-CBa or 2-HBa. In screening tests such as these, it is important to distinguish between potential competitors that are metabolized and those that are not, and in the case of the former, to consider other mechanisms that may give results that emulate competition (recognition by the carrier). These mechanisms include transformation to metabolic inhibitors that interfere with catabolic enzymes or some other aspect of energy metabolism or transformation to the same intermediate(s) as the 14C tracer, which may result in isotope dilution (27). In our estimation, these alternate mechanisms were not significant concerns with either 3-CBa or 2-FBa. The latter was a substrate for the o-halobenzoate-1,2-dioxygenase, and thus, isotope dilution resulting from formation of catechol (and downstream metabolites) could occur. However, 2-FBa was oxidized at a lower rate than either 2-CBa or 2-HBa (13), and if isotope dilution were the only process operative, the level of inhibition caused by 2-FBa should be less, not more, than that of 2-CBa or 2-HBa. 3-Chlorobenzoate could potentially be transformed to 3-chlorocatechol and subsequently to an inhibitory acylhalide. However, based on data from heterologous expression studies and activity exhibited by whole cells (14, 29), no substantial metabolism of 3-CBa was expected by 2-CBa-grown cells of strain D1. As such, we believe that the strong inhibition by 2-FBa and 3-CBa probably reflected preferential recognition by the carrier.
The patterns of substrate recognition by the carrier provided additional proof that uptake of 2-CBa was mediated by a transporter and was not driven by metabolic drag, and they gave insight into the possible mechanisms of carrier-substrate interaction and the substrate range of the carrier. The data were inconsistent with metabolic drag in that the strongest apparent competitor for uptake (3-CBa) was a relatively poor substrate for the o-halobenzoate-1,2-dioxygenase initiating 2-CBa metabolism (14), and conversely, 2,5-diCBa, which is a good substrate for the dioxygenase (14), was not a competitor for uptake. Nonrecognition of 2,5-diCBa, 2,3-diCBa, 2,3,5-triCBa, benzoate, and para-substituted benzoates may have indicated that a single ortho- or meta-substituent was required for substrate binding. The exclusion of these di- and tri-CBas as substrates for the 2-CBa transporter was notable in that metabolism by strain D1 of all of these growth substrates was presumably initiated by the same o-halobenzoate-1,2-dioxygenase (14). Thus, metabolism of these compounds may involve a different transporter or perhaps may be driven by metabolic drag.
Transporter energetics: Artificially imposed electrochemical gradients.
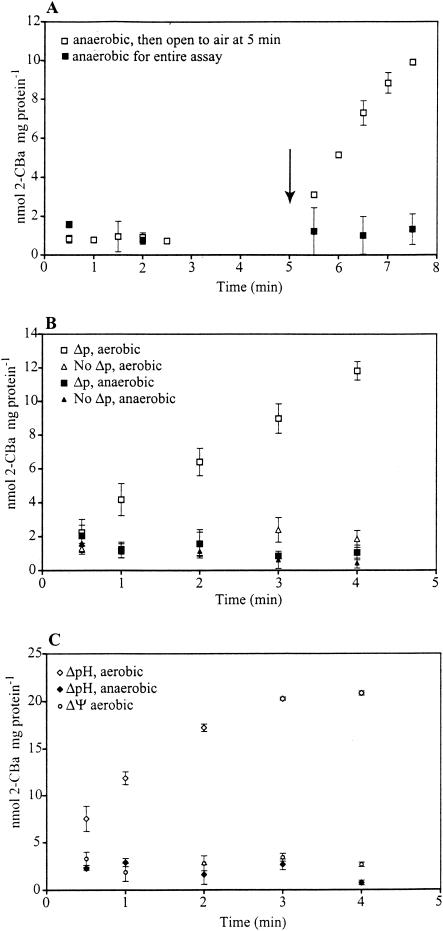
Uptake of 2-CBa was abolished by anaerobic incubation but was rapidly reestablished following exposure to air (Fig. 2A). Amounts of ATP extracted from cells incubated aerobically and anaerobically were 6.11 ± 0.08 and 0.46 ± 0.10 nmol mg of cell protein−1, respectively. Cells treated with valinomycin and K-acetate to dissipate ΔΨ and ΔpH had no detectable 2-CBa uptake when incubated under aerobic or anaerobic conditions (Fig. 2B). For cells diluted in buffers to restore both ΔΨ and ΔpH, or ΔpH alone, uptake of 2-CBa occurred under aerobic but not anaerobic conditions (Figs. 2B and C). In contrast, treatment to restore ΔΨ alone did not reestablish 2-CBa uptake by cells incubated aerobically or anaerobically (Fig. 2C). We attempted to measure the ATP content of cells treated to impose or dissipate gradients, but the luminometer readings were unreliable, apparently because of interference from components of the solutions used.
FIG. 2.
Effect of anaerobic conditions and artificially induced electrochemical gradients on 2-CBa uptake. (A) Cells incubated under anaerobic conditions; the arrow indicates the point at which some selected vials were opened to air. (B) Cells with Δp dissipated or reestablished, incubated under aerobic or anaerobic conditions. (C) Dissipation or reestablishment of ΔpH or ΔΨ in cells incubated under aerobic or anaerobic conditions. Each data point in all figures is the mean value of three replicate assays, and error bars indicate the standard deviation of the means (obscured by symbols in some cases). Cells used in the experiment were harvested from a continuous culture growing on 2-CBa.
The results of these experiments were consistent with those anticipated for an uptake process energized by ATP hydrolysis and not by Δp alone. If the latter were the case, we would have expected 2-CBa uptake by cells incubated anaerobically with Δp restored but uncoupled from ATP formation. An artificial Δp was sufficient to support uptake of 4-chlorobenzoate by the coryneform NTB-1 (10), 4-toluene sulfonate by Comamonas testosteroni T-2 (25), 4-HBa by K. pneumoniae MAO4 (5), and 4-HBa by K. planticola DSZ1 (3). Linkage of 2-CBa uptake to ATP hydrolysis was also indicated by the restoration of activity under aerobic conditions following treatment to reestablish ΔpH but not treatment to reestablish ΔΨ. Allende et al. (2) reported that aerobic incubation and ΔpH supported 4-HBa uptake by Acinetobacter sp. strain BEM2; the dependence of uptake on ATP hydrolysis was further indicated by its elimination following exposure to DCCD.
Transporter energetics: metabolic inhibitors.
The addition of the phosphate analogs vanadate and arsenate had no effect on rates of 2-CBa transport or ATP levels when tested at 1 and 20 mM, respectively (Table 2). However, 50 and 100 mM arsenate significantly decreased (P < 0.01) both uptake and ATP levels. Strain D1 appeared to be more resistant to arsenate than were bacteria examined in other transporter studies where lower concentrations of arsenate effected greater reductions in cellular ATP pools (2, 3, 5, 25). Treatment with 0.5 mM DCCD had no significant effect (P < 0.01) on either uptake rates or cellular ATP pools (Table 2). This compound binds to the F1/Fo ATP synthase (ATPase) complex, preventing ATP synthesis, and the level tested here greatly exceeded that reported to affect ATP synthesis for other bacteria. For example, 5 μM DCCD decreased ATP levels by 92% for Acinetobacter sp. (5), while less than 100 μM DCCD significantly decreased ATP levels for Klebsiella (3, 5), Comamonas (25), Rhizobium (6), and Sphingomonas (38). For strain D1, exposure to KCN, which interrupts electron transfer, nearly eliminated 2-CBa uptake and almost exhausted cellular ATP pools. The protonophores CCCP and 2,4-dinitrophenol, which dissipate ΔΨ and ΔpH, significantly decreased (P < 0.01) uptake rates and ATP levels. Valinomycin, which collapses ΔΨ by rendering cells permeable to K+, had no significant effect (P < 0.01) on either transport or ATP levels. The ionophore nigericin, which dissipates ΔpH by facilitating the exchange of H+ for K+, also did not significantly decrease either uptake rates or ATP levels (Table 2).
TABLE 2.
Effect of metabolic inhibitors on 2-CBa uptake rates and cellular ATP levels measured with 2-CBa-grown cells of strain D1
| Inhibitora | Concn (mM) | Uptake rate (%)b | ATP level (%)c |
|---|---|---|---|
| None | 0 | 100 | 100 |
| Nigericin | 0.025 | 122.8 ± 8.7 | 108.0 ± 3.4 |
| Valinomycin | 0.025 | 102.7 ± 10.5 | 115.4 ± 2.8 |
| DCCD | 0.5 | 106.3 ± 7.4 | 87.8 ± 13.5 |
| VO4−3 | 1 | 100.9 ± 27.2 | 108.3 ± 27.9 |
| AsO4−3 | 20 | 96.3 ± 4.8 | 81.3 ± 15.6 |
| AsO4−3 | 50 | 56.4 ± 2.5*d | 36.1 ± 5.2* |
| AsO4−3 | 100 | 46.9 ± 3.9* | 23 ± 8.1* |
| CCCP | 0.02 | 22.3 ± 5.8* | 47.2 ± 2.1* |
| DNP | 1 | 1.9 ± 2.6* | 19.6 ± 2.1* |
| KCN | 10 | 2.1 ± 3.9* | 6.9 ± 0.1* |
Abbreviations: DCCD, N,N-dicyclohexylcarbodiimide; CCCP, carbonyl cyanide m-chlorophenylhydrazone; DNP, 2,4-dinitrophenol.
100% = 5.5 ± 0.6 nmol min−1 mg of protein−1.
100% = 4.8 ± 0.3 nmol mg of protein−1.
*, values significantly different than the control lacking an inhibitor (P < 0.01).
The strongest trend from the metabolic inhibitor data was that a significant reduction in the 2-CBa uptake rate was accompanied by a significant reduction in the cellular ATP level. This correlation between uptake and cellular ATP levels would be consistent with an ATP-dependent transport process. However, interpretation of the metabolic inhibitor data was complicated by the fact that KCN and protonophores, which had the greatest impact on 2-CBa uptake and the depletion of cellular ATP levels, have multiple physiological effects. Their action in dissipating Δp, which inhibits secondary transporters, is confounded by the accompanying depletion of ATP, which inhibits primary transporters (18, 19, 30, 37).
Role of hybEFG in transport of 2-CBa or 2-HBa.
Mutagenesis experiments were done to determine if hybEFG encoded the transporter that mediated uptake of 2-HBa and 2-CBa. The vector used, pBSL202, contained a mini-Tn5 to effect random insertional mutagenesis (1) and was maintained in E. coli S17-1 λpir. The vector was introduced into strain D1 by electroporation, and a number of gentamicin-resistant, putative strain D1 mutants were recovered. However, hybridization and plasmid screening showed that none of these had transposon inserts, but instead they had retained pBSL202 as a stably replicating plasmid. This was surprising, since replication of the vector is dependent on expression of the pir gene, which is supplied in trans in E. coli and presumably is absent in the recipient (1).
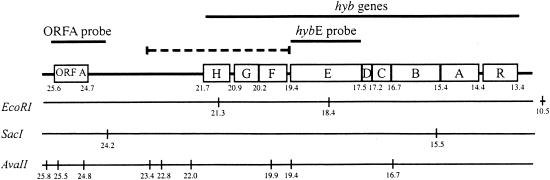
Although the procedure did not generate the intended insertional mutants, PCR screening of the transformants identified one from which hybEFGH failed to amplify. In Southern hybridization analysis of this transformant, a probe to hybE detected a 7.9-kb EcoRI fragment, as expected, but not a second expected 2.9-kb fragment (Fig. 3). In SacI digests of the transformant, the hybE probe hybridized to a 5-kb fragment, which was 3.7 kb smaller than expected (Fig. 3). Probing AvaII digests with hybE yielded the expected 2.7-kb fragment (Fig. 3). Hybridization of an ORFA probe to AvaII-digested DNA gave the expected fragments, including the 1.4-kb fragment that was bounded by map positions 24.8 and 23.4 kb (Fig. 3). Collectively, the hybridization patterns of hybE in the EcoRI, SacI, and AvaII digests suggested that hybE (and the upstream hyb region) was intact but that a ca. 3.7-kb deletion occurred downstream of hybE. The exact points of deletion were not determined, but we could conclude that it occurred within the 4-kb region bounded downstream by map position 23.4 kb and upstream by map position 19.4 (Fig. 3). This deletion effectively eliminated hybF and hybG, the putative products of which had significant identity to ATP-binding proteins of an ABC-type transporter.
FIG. 3.
Physical map of the hybEFG genes and adjoining regions. Locations of restriction sites for the enzymes listed are indicated by the vertical bars and are numbered (in kilobases) relative to distance from the beginning of the 26-kb region in which they were originally identified (15). For EcoRI, a restriction site located at 10.5 kb that is off the map at the scale used is indicated by the gap. The dashed line indicates the 4-kb region in which the 3.7-kb deletion occurred in the hybFG mutant. Solid lines indicate probes used in Southern hybridization analysis of the mutant.
The deletion mutant retained the ability to grow on 2-CBa and 2-HBa, and its uptake of these compounds was examined. For batch-grown cells cultured on 2-CBa, the average 2-[14C]CBa uptake rates for the mutant and wild type were 0.69 ± 0.38 and 0.85 ± 0.46 nmol min−1 mg of protein−1, respectively. The average 2-[14C]HBa uptake rates were 0.85 ± 0.34 and 0.79 ± 0.30 nmol min−1 mg of protein−1 for the mutant and wild type. For both 2-CBa and 2-HBa, the uptake rates by mutant and wild-type cells were not significantly different (P < 0.01). These levels of transporter activity are lower than those reported above for 2-CBa/2-HBa-grown batch-culture cells; this difference reflected the batch-to-batch variability observed in the uptake assays.
The activity of ABC transporters is dependent on the hydrolysis of ATP, which is mediated by specific ATP-binding proteins with which they associate (16). Thus, the loss of the hybF and hybG would be expected to render the putative transporter encoded by hybEFG nonfunctional. Since 2-CBa or 2-HBa uptake by a deletion mutant was no different from that of the wild type, it was unlikely that a hybEFG-encoded transporter mediated this process. However, the fact that strain D showed no uptake of 2-CBa suggests that genes encoding the transporter were carried on the same mobile element as the hyb and ohb genes.
Conclusions.
Uptake of 2-CBa and 2-HBa is mediated by an inducible transporter in strain D1. The relative rates of activity for the transporter were comparable with those reported for other aromatic acids. The substrate range of the transporter did not include other o-halobenzoates that serve as growth substrates for strain D1 and for which the metabolism is initiated by the same dioxygenase as for 2-CBa and 2-HBa. This suggests that multiple mechanisms for substrate uptake were coupled to the same catabolic enzyme. Substrate binding and/or translocation appeared to be favored by a single, electronegative substituent in the ortho or meta position. The preponderance of evidence suggested that 2-CBa uptake was driven by ATP hydrolysis. If so, the 2-CBa transporter would be the first of the ABC type implicated in uptake of haloaromatic acids.
Acknowledgments
We thank M. F. Alexeyev for the kind gift of the pBSL202 vector. We acknowledge and thank Vera Jencova for doing the chemostat washout experiments.
These studies were funded by U.S. EPA grant R82-7103-01-0 to W.J.H. and Hatch project W1504466 to W.J.H.
REFERENCES
- 1.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. New mini-Tn5 derivatives for insertion mutagenesis and genetic engineering in Gram-negative bacteria. Can. J. Microbiol. 41:1053-1055. [DOI] [PubMed] [Google Scholar]
- 2.Allende, J. L., A. Gibello, A. Fortun, G. Mengs, E. Ferrer, and M. Martin. 2000. 4-Hydroxybenzoate uptake in an isolated soil Acinetobacter sp. Curr. Microbiol. 40:34-39. [DOI] [PubMed] [Google Scholar]
- 3.Allende, J. L., A. Gibello, A. Fortun, M. Sanchez, and M. Martin. 2002. 4-Hydroxybenzoate uptake in Klebsiella planticola DSZ1 is driven by delta pH. Curr. Microbiol. 44:31-37. [DOI] [PubMed] [Google Scholar]
- 4.Allende, J. L., A. Gibello, M. Martin, and A. Garrido-Pertierra. 1992. Transport of 4-hydroxyphenylacetic acid in Klebsiella pneumoniae. Arch. Biochem. Biophys. 292:583-588. [DOI] [PubMed] [Google Scholar]
- 5.Allende, J. L., M. Suarez, M. Gallego, and A. Garrido-Pertierra. 1993. 4-Hydroxybenzoate uptake in Klebsiella pneumoniae is driven by electrical potential. Arch. Biochem. Biophys. 300:142-147. [DOI] [PubMed] [Google Scholar]
- 6.Botero, L. M., T. S. Al-Niemi, and T. R. McDermott. 2000. Characterization of two inducible phosphate transport systems in Rhizobium tropici. Appl. Environ. Microbiol. 66:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye. Anal. Biochem. 72:248-253. [DOI] [PubMed] [Google Scholar]
- 8.Chang, H.-K., and G. J. Zylstra. 1999. Characterization of the phthalate permease OphD from Burkholderia cepacia ATCC 17616. J. Bacteriol. 181:6197-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collier, L. S., N. N. Nichols, and E. L. Neidle. 1997. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J. Bacteriol. 179:5943-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groenewegen, P. E. J., A. J. M. Driessen, W. N. Konings, and J. A. M. D. Bont. 1990. Energy-dependent uptake of 4-chlorobenzoate in the coryneform bacterium NTB-1. J. Bacteriol. 172:419-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harwood, C. S., and J. Gibson. 1986. Uptake of benzoate by Rhodopseudomonas palustris grown anaerobically in light. J. Bacteriol. 165:504-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harwood, C. S., N. N. Nichols, M.-K. Kim, J. L. Ditty, and R. E. Parales. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickey, W. J., and D. D. Focht. 1990. Degradation of mono-, di-, and trihalogenated benzoic acids by Pseudomonas aeruginosa JB2. Appl. Environ. Microbiol. 56:3842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hickey, W. J., and G. Sabat. 2001. Integration of matrix-assisted laser desorption ionization time-of-flight mass spectrometry and molecular cloning for the identification and functional characterization of mobile ortho-halobenzoate oxygenase genes in Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:5648-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hickey, W. J., G. Sabat, A. S. Yuroff, A. A. Arment, and J. Perez-Lesher. 2001. Cloning, nucleotide sequencing, and functional analysis of a novel, mobile cluster of biodegradation genes from Pseudomonas aeruginosa JB2. Appl. Environ. Microbiol. 67:4603-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, S. J., and J. Mandelstam. 1972. Evidence for induced synthesis of an active transport factor for mandelate in Pseudomonas putida. Biochem. J. 126:917-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs, M. H. J., A. J. M. Driessen, and W. N. Konings. 1995. Characterization of a binding protein-dependent glutamate transport system of Rhodobacter sphaeroides. J. Bacteriol. 177:1812-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi, A. K., S. Ahmed, and G. F.-L. Ames. 1989. Energy coupling in bacterial periplasmic transport systems. J. Biol. Chem. 264:2126-2133. [PubMed] [Google Scholar]
- 20.Kashet, E. R. 1985. The proton motive force in bacteria: a critical assessment of methods. Annu. Rev. Microbiol. 39:219-342. [DOI] [PubMed] [Google Scholar]
- 21.Krooneman, J., E. R. B. Moore, J. C. L. van Velzen, R. A. Prins, L. J. Forney, and J. C. Gottschal. 1998. Competition for oxygen and 3-chlorobenzoate between two aerobic bacteria using different degradation pathways. FEMS Microbiol. Ecol. 26:171-179. [Google Scholar]
- 22.Krooneman, J., A. O. Sliekers, T. M. P. Gomes, L. J. Forney, and J. C. Gottschal. 2000. Characterization of 3-chlorobenzoate degrading aerobic bacteria isolated under various environmental conditions. FEMS Microbiol. Ecol. 32:53-59. [DOI] [PubMed] [Google Scholar]
- 23.Krooneman, J., S. Van Den Akker, T. M. P. Gomes, L. J. Forney, and J. C. Gottschal. 1999. Degradation of 3-chlorobenzoate under low-oxygen conditions in pure and mixed cultures of the anoxygenic photoheterotroph Rhodopseudomonas palustris DCP3 and an aerobic Alcaligenes species. Appl. Environ. Microbiol. 65:131-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leveau, J. H. L., A. J. B. Zehnder, and J. R. van der Meer. 1998. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 180:2237-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locher, H. H., B. Poolman, A. M. Cook, and W. N. Konings. 1993. Uptake of 4-toluene sulfonate by Comamonas testosteroni T-2. J. Bacteriol. 175:1075-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkel, S. M., A. E. Eberhard, J. Gibson, and C. S. Harwood. 1989. Involvement of coenzyme A thioesters in anaerobic metabolism of 4-hydroxybenzoate by Rhodopseudomonas palustris. J. Bacteriol. 171:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miguez, C. B., C. W. Greer, J. M. Inggram, and R. A. MacLeod. 1995. Uptake of benzoic acid and chloro-substituted benzoic acids by Alcaligenes denitrificans BRI 3010 and BRI 6011. Appl. Environ. Microbiol. 61:4152-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols, N. N., and C. S. Harwood. 1997. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Lesher, J., and W. J. Hickey. 1995. Use of an s-triazine nitrogen source to select for and isolate a recombinant chlorobenzoate-degrading Pseudomonas. FEMS Microbiol. Lett. 133:47-52. [Google Scholar]
- 30.Poolman, B. 1999. Regulation of solute accumulation in bacteria and its physiological significance, p. 41-57. In J. K. Broome-Smith, S. Baumberg, C. J. Stirling, and F. B. Ward (ed.), Transport of molecules across microbial membranes. Cambridge University Press, Cambridge, United Kingdom.
- 31.Prieto, M. A., and J. L. Garcia. 1997. Identification of the 4-hydroxyphenylacetate transport gene of Escherichia coli W: construction of a highly sensitive cellular biosensor. FEBS Lett. 414:293-297. [DOI] [PubMed] [Google Scholar]
- 32.Saint, C. P., and P. Romas. 1996. 4-Methylphthalate catabolism in Burkholderia cepacia Pc701: a gene encoding a phthalate-specific permease forms part of a novel gene cluster. Microbiology 142:2407-2418. [DOI] [PubMed] [Google Scholar]
- 33.Schleissner, C., E. R. Olivera, M. Fernandez-Valverde, and J. M. Luengo. 1994. Aerobic catabolism of phenylacetic acid in Pseudomonas putida U: biochemical characterization of a specific phenylacetic acid transport system and formal demonstration that phenylacetyl-coenzyme A is a catabolic intermediate. J. Bacteriol. 176:7667-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaler, T. A., and G. M. Klecka. 1986. Effects of dissolved-oxygen concentration on biodegradation of 2,4-dichlorophenoxyacetic acid. Appl. Environ. Microbiol. 51:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thayer, J. R., and M. L. Wheelis. 1982. Active transport of benzoate in Pseudomonas putida. J. Gen. Microbiol. 128:1749-1753. [DOI] [PubMed] [Google Scholar]
- 36.van der Woude, B. J., J. C. Gottschal, and R. A. Prins. 1995. Degradation of 2,5-dichlorobenzoic acid by Pseudomonas aeruginosa JB2 at low oxygen tension. Biodegradation 6:39-46. [DOI] [PubMed] [Google Scholar]
- 37.Wong, C. M., M. J. Dilworth, and A. R. Glenn. 1994. Cloning and sequencing show that 4-hydroxybenzoate hydroxylase (PobA) is required for uptake of 4-hydroxybenzoate in Rhizobium leguminosarum. Microbiology 140:2775-2786. [DOI] [PubMed] [Google Scholar]
- 38.Zipper, C., M. Bunk, A. J. B. Zehnder, and H.-P. E. Kohler. 1998. Enantioselective uptake and degradation of chiral herbicide dichlorprop [(RS)-2-(2, 4-dichlorophenoxy)propanoic acid] by Sphingomonas herbicidovorans MH. J. Bacteriol. 180:3368-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]