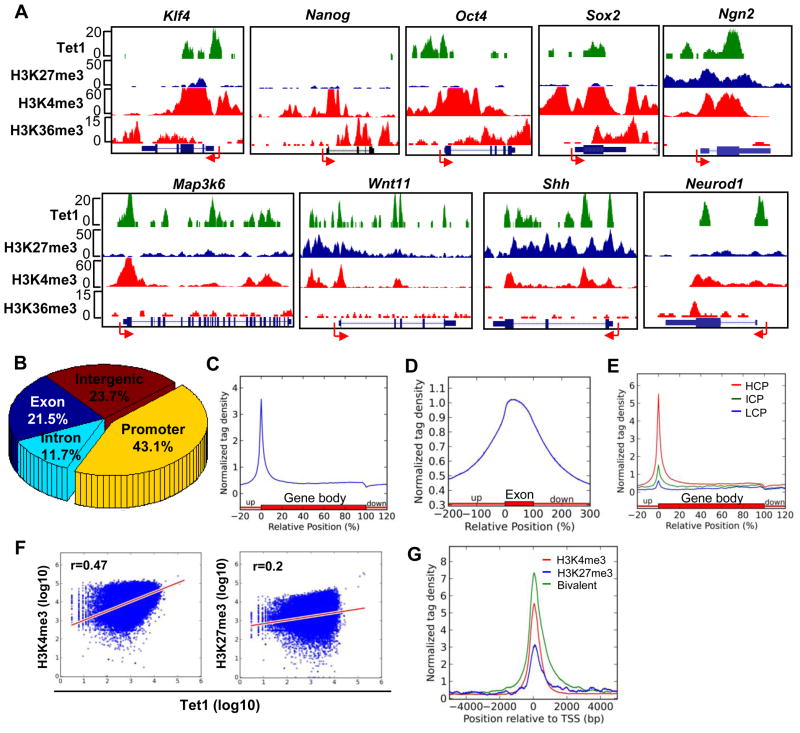
Figure 2. Tet1 is enriched at gene promoters and positively correlates with promoter CpG content and H3K4me3.
(A) Representative Tet1 ChIP-seq results and associated histone modification patterns. Arrow denotes promoter orientation.
(B) Genomic distribution of Tet1-enriched regions. The genomic features (exons, introns, and intergenic regions) were defined based on RefSeq gene (mm9) annotations. Promoter was defined as −2kb to +2kb relative to TSS.
(C) Normalized Tet1 tag density distribution across the gene body. Each gene body was normalized to 0–100%. Normalized Tag density is plotted from 20% of upstream of TSSs to 20% downstream of TTSs.
(D) Normalized Tet1 tag density distribution across exons. Each exon is normalized to 0–100%. Normalized Tet1 density is plotted from 200% upstream to 200% downstream of the normalized exon.
(E) Normalized Tet1 tag density distribution across gene bodies with promoter classifications. The high CpG promoters (HCPs), intermediate CpG promoters (ICPs) and low CpG promoters (LCPs) were defined as described previously (Meissner et al., 2008; Mikkelsen et al., 2007). Each gene body region was normalized to 0–100%. Normalized tag density is shown from 20% upstream of TSSs to 20% downstream of TTSs.
(F) The correlations between Tet1 and H3K4me3 (left) or H3K27me3 (right) tag densities at gene promoters. H3K4me3 and H3K27me3 densities were obtained from the reference (Mikkelsen et al., 2007). Tet1, H3K4me3, and H3K27me3 values at promoters were calculated as log10 (tag density).
(G) Normalized Tet1 tag density distributions at ‘univalent’ H3K4me3 (red), ‘univalent’ H3K27me3 (blue) and ‘bivalent’ (green) promoters. Promoters were classified according to their histone modification patterns (Mikkelsen et al., 2007). Tet1 enrichment from −5kb to +5kb relative to TSSs is shown.