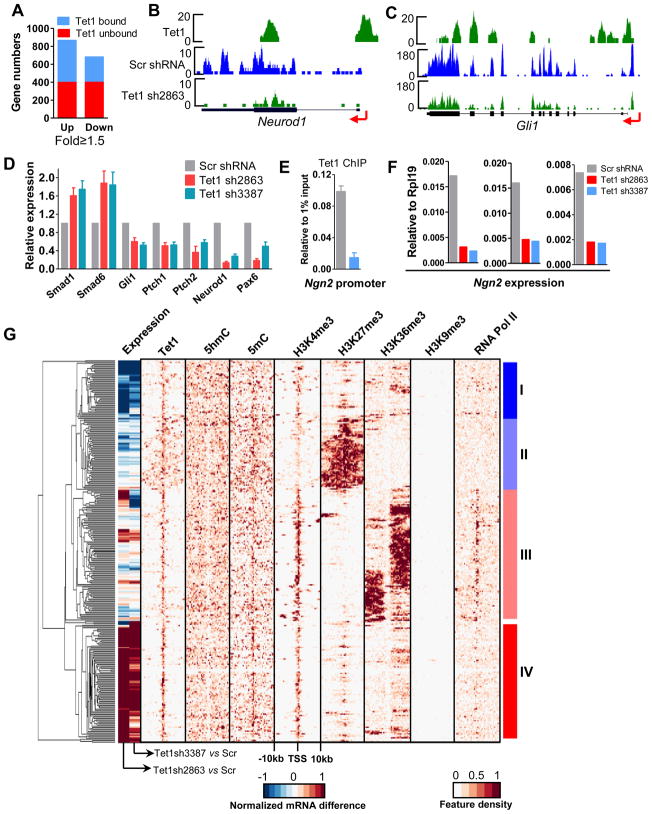
Figure 6. Tet1 plays both positive and negative roles in target gene regulation in mESCs.
(A) Using 1.5-fold as a cut-off value, all differentially expressed genes in the microarray assay were selected and further classified into Tet1 bound and unbound genes.
(B–C) Representative mRNA-seq results. Arrow denotes promoter orientation.
(D) RT-qPCR confirms the differentially expressed genes after Tet1 depletion. Results are shown as mean +/− SEM (n=4).
(E) Tet1 ChIP-qPCR reveals that Tet1 binds to the Ngn2 promoter and Tet1 depletion causes the significantly decreased Tet1 occupancy at the Ngn2 promoter in mESCs. Data are presented as mean +/− SEM (n=3).
(F) Tet1 depletion delayed the Ngn2 induction during neural differentiation. ESCs were infected with control or Tet1 shRNA containing lentivirus, selected with puromycin and differentiated by LIF withdraw for 3 days. The formed embryonic bodies were treated with 1μM RA for 3 days. Ngn2 expression was examined by RT-qPCR. Results of three independent experiments are shown.
(G) Hierarchical clustering of epigenetic and transcriptional features of Tet1 target genes. The histone methylation, DNA methylation and RNA Pol II profiles in mESCs were obtained from the references (Meissner et al., 2008; Mikkelsen et al., 2007). Top 300 Tet1 target genes based on the variance of expression and epigenetic modifications density score were chosen to make the cluster. The tag densities of Tet1 binding, 5hmC, 5mC, H3K4me3, H3K27me3, H3k36me3, H3K9me3 and RNA Pol II were profiled through −10kb to +10kb relative to the TSS of each gene with bin of 500bp. The middle color bar indicates the difference of mRNA levels between Scr shRNA- and Tet1 shRNA2863- or Tet1 shRNA3387-treated mESCs.