Abstract
The orphan G protein-coupled receptor GPR124/TEM5 is highly expressed in central nervous system (CNS) endothelium. Here, complete null or endothelial-specific GPR124 deletion produced embryonic lethality from CNS-specific angiogenesis arrest in forebrain and neural tube. Conversely, GPR124 overexpression throughout all adult vascular beds produced CNS-specific hyperproliferative vascular malformations. In vivo, GPR124 functioned cell-autonomously in endothelium to regulate sprouting, migration, and developmental expression of the blood-brain barrier marker Glut1, while in vitro, GPR124 mediated Cdc42-dependent directional migration to forebrain-derived, VEGF-independent cues. Our results demonstrate CNS-specific angiogenesis regulation by an endothelial receptor, and illuminate functions of the poorly understood adhesion GPCR subfamily. Further, the striking functional tropism of GPR124 marks this receptor as a therapeutic target for CNS-related vascular pathologies.
Organ-specific vascular beds form by angiogenesis, followed by significant anatomic and molecular differentiation to support various physiologic requirements (1, 2). The central nervous system (CNS) vasculature is highly specialized, with a blood-brain barrier (BBB), extensive pericyte coverage, reciprocal interactions with neurons and glia, and function as a neural stem cell niche (3–5). In the developing CNS, the perineural vascular plexus (PNVP) surrounds the ventral neural tube at E7.5-8.5. By E 11.5, angiogenic endothelial sprouts invade the neuroepithelium from the pial surface to periventricular areas and branch, generating the periventricular vascular plexus (PVVP) (6, 7). This angiogenic invasion has been proposed to occur concomitantly with barriergenesis, the acquisition of BBB properties (8, 9). Heretofore, endothelial receptors mediating CNS-specific angiogenesis have not been described.
We investigated GPR124/Tumor Endothelial Marker 5 (TEM5), which is an orphan G-protein coupled receptor (GPCR) with a large ectodomain belonging to the adhesion GPCR subfamily (fig. S1A). GPR124/TEM5 was initially identified as differentially expressed in tumor vasculature (10, 11) and in genomic analysis of GPCR loci (12). During mouse embryogenesis, GPR124 was expressed in both endothelial cells and pericytes, most prominently in brain and neural tube (fig. S1, B to L), and to lesser degrees in non-CNS embryonic organs including liver, heart and kidney (fig S2). GPR124 was also expressed in embryonic epithelium of lung and esophagus and in mesenchyme (fig. S2). In contrast, adult GPR124 expression was exclusively vascular in tissues examined, with endothelial expression in CNS including brain and retina (fig. S3) and more widespread pericyte expression in brain and organs including kidney, pancreas and corpus luteum (fig. S4).
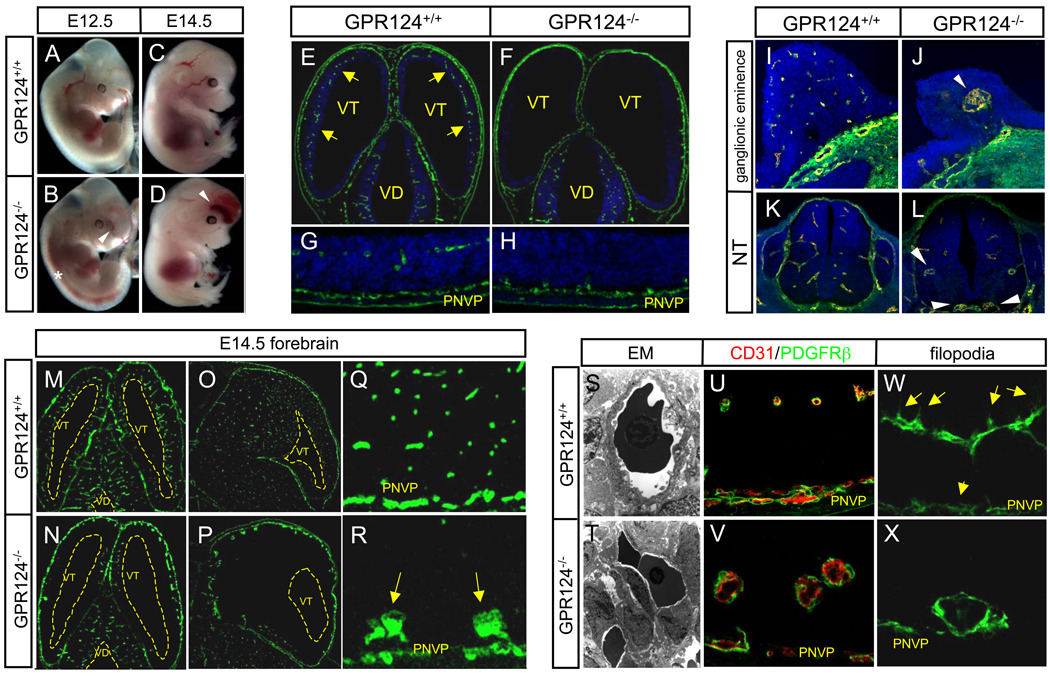
To examine the function of this orphan receptor, we generated a GPR124 lacZ knock-in null allele, which displayed a vascular embryonic expression pattern upon β-galactosidase staining of heterozygous embryos at E10.5 (figs. S5, S6). No viable adult GPR124−/− (i.e. GPR124lacZ/lacZ) mice were obtained (fig. S7). Beginning at E11, GPR124−/− embryos exhibited completely penetrant, progressive CNS hemorrhage originating in forebrain telencephalon and ventral neural tube (Fig. 1, A to D, and figs. S7 and S8), with resultant embryonic lethality by E15.5 (fig. S7). Paralleling sites of hemorrhage, GPR124−/− embryos at E11.5 displayed selective CNS-specific vascular patterning defects, with markedly reduced angiogenic sprouting into the forebrain telencephalon and significant thickening of the underlying PNVP, rendering the telencephalon virtually avascular (Fig. 1, E to H). By E14.5, this culminated in formation of basally localized glomeruloid malformations and resultant distally unvascularized telencephalon in GPR124−/− embryos. In contrast, wild type mice showed efficient migration of endothelial cells to the subventricular zone (SVZ) (Fig. 1, M to R). Similar striking angiogenic deficits with characteristic hemorrhagic glomeruloid malformations were also seen in GPR124−/− ventral neural tube and forebrain ganglionic eminences (Fig. 1, I to L). Notably, GPR124 deletion did not impair angiogenic sprouting into the embryonic diencephalon, mid- and hindbrain, or the vascularization of non-neural tissues (Fig 1. E to F, M to N, fig. S9 and S10). Electron microscopy of GPR124−/− CNS vascular malformations revealed numerous packed endothelial cells with scant cytoplasm, enveloped by PDGFRβ+ pericytes (Fig 1, S to V). Further, GPR124−/− endothelium showed markedly reduced to absent filopodial extensions which when present extended laterally rather than towards the ventricles as in wildtype (Fig. 1, W to X). Proliferation, apoptosis and VEGF receptor expression were grossly unaltered in GPR124−/− vascular malformations (fig. S11). We conclude that GPR124 displays a pronounced functional tropism for the CNS and is essential for developmental angiogenic sprouting into forebrain and neural tube.
Fig. 1.
(A to X) CNS vascular patterning defects in GPR124−/− embryos. GPR124−/− embryos manifested progressive hemorrhage confined to forebrain telencephalon (arrowheads) and neural tube (*) [(A) to (D)]. Wild type E11.5 embryos exhibited efficient sprouting angiogenesis (arrows) from the perineural vascular plexus (PNVP) into the telencephalon [(E) and (G)], which was completely ablated in GPR124−/− embryos that exhibited PNVP thickening only [(F) to (H)], Green- CD31 IF. Large, glomeruloid vascular malformations (arrowheads) and absence of a vascular network were observed in E11.5 GPR124−/− forebrain ganglionic eminences and ventral neural tube [(I) to (L), CD31 IF]. By E14.5, GPR124−/− telencephalon was essentially devoid of a mature capillary network with glomeruloid malformations abutting the pia/PNVP, while the diencephalon was unaffected [(M) to (N), transverse; (O) to (P), sagittal; vascular malformations (arrows), (Q) and (R), CD31 IF]. Glomeruloid malformations were irregular, multi-layer endothelial aggregates versus single layer capillaries in controls [(S) and (T), EM 4300x], with normal peripheral PDGFRβ+ pericyte investment [(U) and (V)] but lacked ventricularly-directed endothelial filopodia (arrows) [(W) and (X), CD31 IF]; VD, ventricle diencephalons; VT, ventricle telencephalon; blue, DAPI.
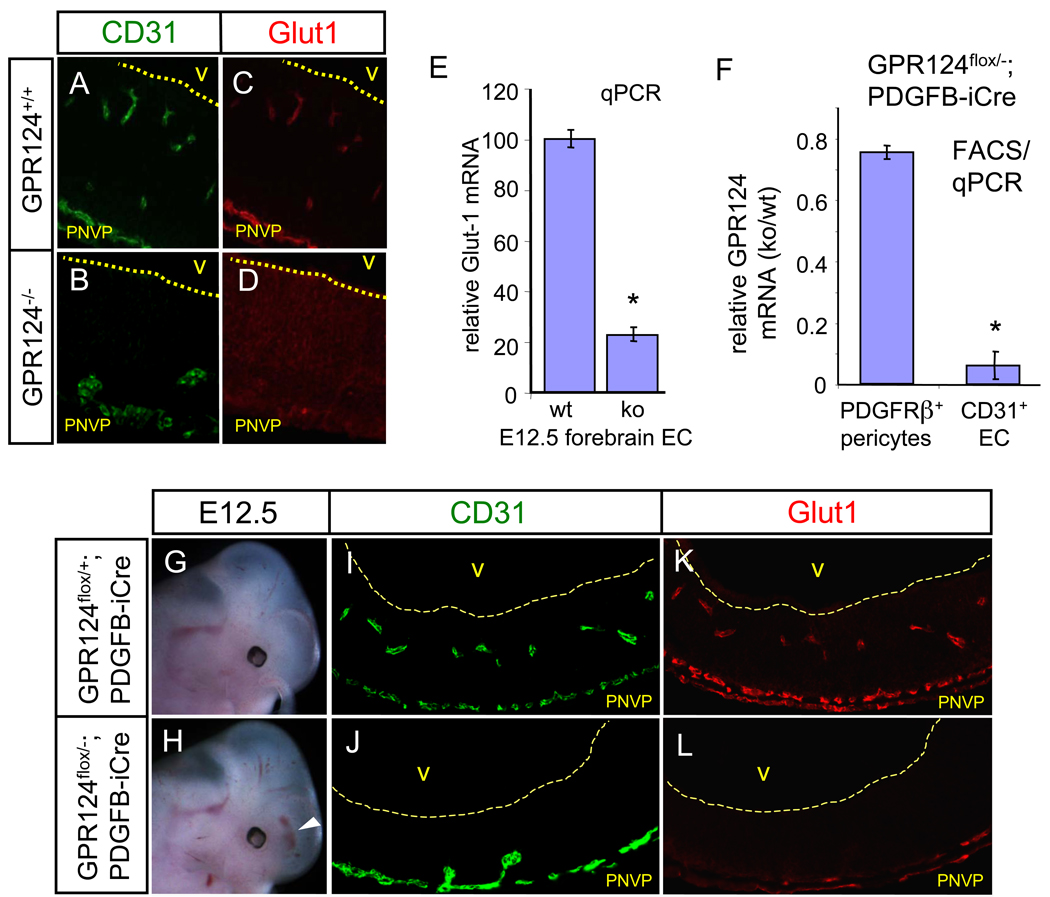
Wnt7/β-catenin knockouts with CNS angiogenesis deficits exhibit defective developmental expression of the BBB-specific glucose transporter Glut1, suggesting that acquisition of BBB markers may be coupled to developmental brain angiogenesis (8, 9). Glut1 expression was similarly absent in GPR124−/− endothelium in forebrain upon immunofluorescence and qPCR analysis of purified primary forebrain GPR124−/− EC (Fig. 2, A to E); expression in mid- and hindbrain was unaffected. Compensatory neuroepithelial Glut-1 upregulation was present (Fig. 2, C and D) as also seen in Wnt7/β-catenin knockouts (8, 9).
Fig. 2.
(A to L) GPR124 functions cell-autonomously in endothelium. Expression of Glut1 is lost in GPR124−/− CNS endothelium, with compensatory Glut1 upregulation in neuroepithelium, [(A) to (D)], E12.5. Glut1 is strongly down-regulated in FACS-isolated, GPR124−/− telencephalic endothelium. qPCR, n=9, E12.5 [(E)]. Selective deletion of GPR124 in FACS-isolated forebrain CD31+ endothelium but not PDGFRβ+ pericytes. qPCR, n=6, E 12.5 [(F)]. E12.5 GPR124flox/−; PDGFB-iCre embryos exhibit forebrain hemorrhaging, formation of PNVP--associated glomeruloid vascular malformations and endothelial Glut1 downregulation [(G) to (L)], recapitulating the global GPR124 deletion phenotype. PNVP, perineural vascular plexus. Error bars are +/− 1 S. D.
To demonstrate endothelial cell-autonomous function of GPR124 in regulating CNS angiogenesis we generated a floxed exon 1 GPR124 allele (GPR124flox) and crossed to PDGFB-iCre mice to allow endothelial CreER-mediated deletion (13). Accordingly, E12.5 GPR124flox/−; PDGFB-iCre embryos demonstrated selective GPR124 deletion in endothelium but not pericytes by FACS/qPCR (Fig. 2F). Notably, GPR124flox/−; PDGFB-iCre mice fully recapitulated the GPR124 null phenotype with forebrain hemorrhage, CNS angiogenic arrest, basally localized glomeruloid malformations and loss of endothelial Glut1 expression (Fig. 2, G to L). Identical results were obtained with an independent vascular deletion strategy in GPR124flox/−; Tie2-Cre mice (fig. S12). These results indicate that GPR124 functions cell-autonomously in CNS endothelium with Glut1 as a developmentally regulated downstream locus.
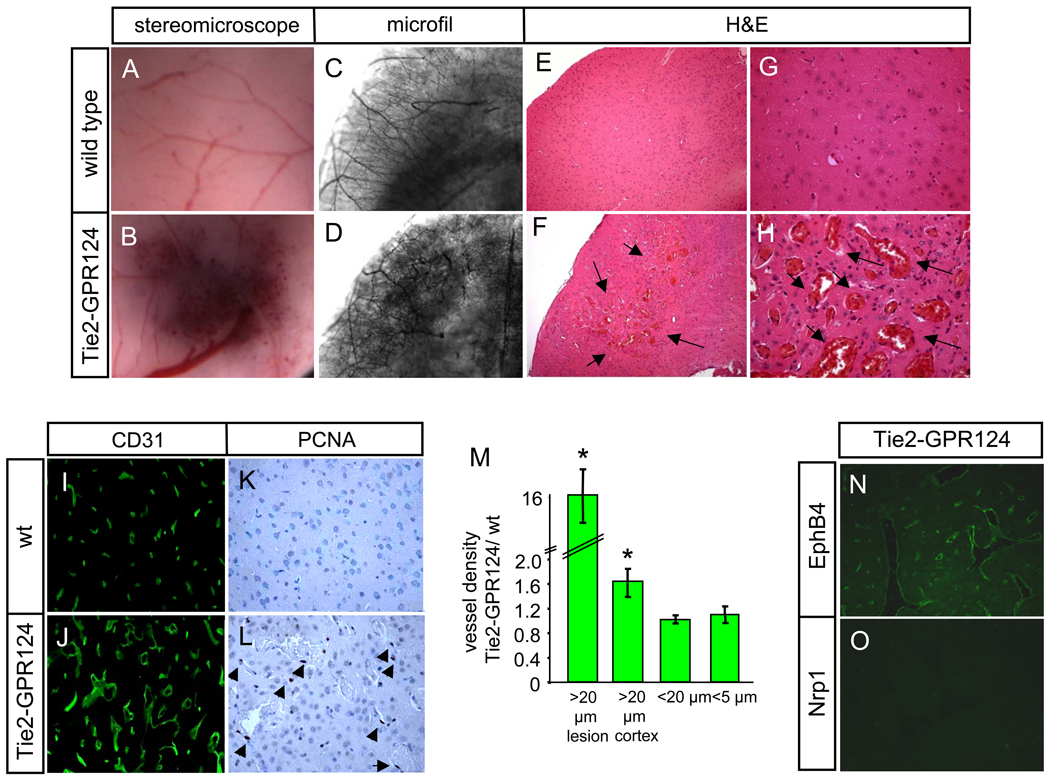
To analyze gain-of-function phenotypic outcomes, we generated transgenic mice overexpressing GPR124 cDNA from the Tie2 promoter/long enhancer active in embryonic and adult endothelium (14). Tie2-GPR124 transgenic mice were viable and fertile. The mutant mice exhibited enhanced brain vascular GPR124 immunofluorescence versus non-transgenic littermates, and displayed ectopic GPR124 expression throughout the microvasculature of adult organs that do not normally express GPR124 such as heart and liver (fig. S13). Despite widespread GPR124 overexpression, Tie2-GPR124 animals displayed a progressive brain-specific vascular phenotype characterized by enlarging oligo-focal areas of hypervascularity that were predominantly cortical (>90%, with ~ 10% in the cerebellum), and presented with ~70% penetrance by 12 months of age, paralleling the forebrain telencephalon distribution of GPR124−/− malformations (Fig. 3, A to H, M); non-CNS organs were unaffected (fig. S14). These lesions consisted of tangled, thin-walled, CD31-positive, grossly dilated hyperproliferative capillaries with associated PDGFRβ+ pericytes (Fig 3, I to L, fig. S15). The vascular malformations exhibited intervening neural tissue reminiscent of venous angiomas and expressed the venous marker EphB4 but not the arterial marker Nrp1 (Fig. 3, N and O). Calcifications were frequently associated (fig. S15E) and transgenic mice occasionally exhibited neurologic symptoms such as ataxia. Thus, both the Tie2-GPR124 overexpressing mice and GPR124−/− embryos displayed angiogenic phenotypes highly restricted to the CNS vasculature.
Fig. 3.
(A to O) CNS vascular malformations in adult (12 months old) Tie2-GPR124 transgenic animals. Focally increased red blood cell accumulation in Tie2-GPR124 cerebral cortex [(A) and (B)] corresponded to increased vascular density and tortuous, enlarged vessels upon microfil vascular casting [(C) and (D)]. H&E staining of a hypervascular area (arrows) in Tie2-GPR124 cortex, 100x [(E) and (F)] revealed dramatically enlarged, engorged thin-walled vessels with intervening neural tissue reminiscent of venous angiomas, 400x [(G) and (H)]. Tie2-GPR124 transgenics exhibited increased CD31+ microvessel density and markedly enlarged [(I) to (J), (M)] and hyperproliferative vascular malformations versus wild type controls [(K) and (L), arrowheads denote PCNA+ cells]. Tie2-GPR124 CNS vascular malformations expressed the venous marker EphB4, but not the arterial receptor Nrp1 [(N) and (O)]. Error bars are +/− 1 S. D.
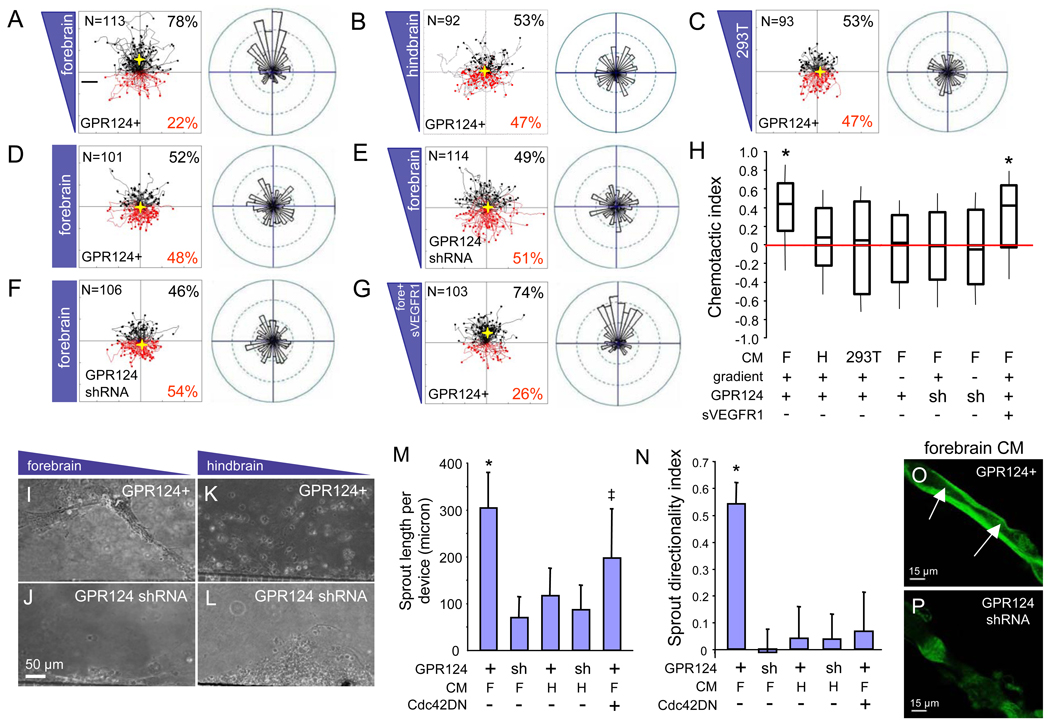
The angiogenic defects in GPR124−/− forebrain but not mid- or hindbrain suggested that GPR124 could mediate endothelial migration towards forebrain-enriched regional guidance cues within the embryonic CNS. The GPR124-expressing brain endothelial cell line bEND3 was used in a microfluidic migration chamber allowing extremely stable, shear-minimized linear chemoattractant gradients and real-time video microscopy of cellular motility (15) (fig. S16A). Notably, GPR124-overexpressing bEND3 cells (GPR124+) (fig. S16B) exhibited directed migration in microfluidic chambers towards gradients of conditioned medium from cultured E12.5 forebrain cortical cells (F), but migrated randomly when presented with equivalent hindbrain preparations (H) or with HEK 293T conditioned medium (chemotactic indices of 0.44, 0.08 and 0.02, respectively) (Fig. 4, A to C, H, and fig. S17). Further, directed migration was not observed when forebrain conditioned medium was evenly distributed across the microfluidic chamber (no gradient), or upon GPR124− shRNA knockdown (Fig. 4, D to F, H, S17). GPR124+ bEND3 migration to the forebrain CM gradient was insensitive to a recombinant soluble VEGFR1 ectodomain, consistent with VEGF-independence. (Fig. 4, G and H, fig. S17).
Fig. 4.
(A to O) GPR124 regulates CNS endothelial migration and sprout formation in vitro. Motility of the GPR124-expressing brain endothelial line bEND3 was monitored by real-time video microscopy in a microfluidic migration chamber allowing extremely stable, shear-minimized linear chemoattractant gradients. GPR124-overexpressing bEND3 cells (GPR124+) exhibited directed migration towards gradients of conditioned medium (CM) from cultured E12.5 forebrain cortical cells, but migrated randomly to equivalent hindbrain or HEK 293T CM [(A) to (C), tracks of individual cells (left) and angular histograms (right) are shown]. No directed migration was observed with even distribution of forebrain CM across the microfluidic chamber (no gradient), or GPR124 shRNA [(D) to (F)]; migration was further insensitive to a recombinant soluble VEGFR1 ectodomain (400 ng/ml), indicating VEGF-independence [(G)]. Box plot of chemotactic indices of bEND3 migration [(H), center line: median, boxes: upper and lower quartiles, *= p<0.001]. In a sprout formation assay GPR124+ bEND3 displayed collective cell migration and robust formation of stable sprouts towards forebrain CM gradients, but not to hindbrain CM nor upon GPR124 shRNA knockdown [(I) to (L), (M)]. Dominant-negative Cdc42N17 inhibited sprout directionality but not elongation [(M) to (N), *= p<0.05, ‡ = N.S.]. GPR124+ sprouts towards forebrain CM contained stable, contiguous lumens, while rare GPR124 shRNA sprouts were unstable and failed to lumenize. Confocal microscopy [(O) to (P)]. F, forebrain CM; H, hindbrain CM. Error bars are +/− 1 S. E. *= p<0.05. N= 3–5 chambers per condition.
We also utilized a second microfluidic assay in which bEND3 cells sprouted through microcapillaries into an acellular chamber containing a transverse gradient of chemoattractant (fig. S18). This revealed multicellular endothelial sprout elongation and directional migration towards forebrain but not hindbrain conditioned medium gradients that was ablated by GPR124 shRNA (Fig. 4, I to N). Cdc42 was required for GPR124-dependent directional migration but not sprout elongation as revealed by a dominant-negative Cdc42 allele (Cdc42N17) (Fig. 4, M to N). Notably, GPR124+ sprouts to forebrain conditioned medium demonstrated contiguous lumen formation upon 3D confocal reconstruction, which was not observed with GPR124 shRNA (Fig. 4, O and P), paralleling lumen enlargement in Tie2-GPR124 transgenics (Fig. 3, H and J). Thus, GPR124 mediates cell-autonomous and VEGF-independent regulation of endothelial migration in response to factor(s) secreted from embryonic forebrain neuroepithelium.
Our studies demonstrate that GPR124/TEM5 is a novel pro-angiogenic, endothelial receptor with pronounced functional tropism for the CNS vasculature in both loss-of-function and gain-of-function settings. Although GPR124 was initially described as upregulated in neoplastic, versus resting endothelium (10, 11), our work establishes its function in normal tissues as a previously unsuspected and essential regulator of CNS angiogenesis, via an endothelial cell-autonomous pathway regulating Cdc42-dependent angiogenic migration and sprouting, Despite the atretic and misoriented filopodia in GPR124 ko vasculature, GPR124-deficient endothelium can incorporate into “tip cell” positions upon mosaic analysis (fig. S19).
Since GPR124 is expressed at all levels of the CNS vasculature, the forebrain-and ventral neural tube-specific angiogenic deficits suggest a corresponding embryonic spatial restriction of either GPR124 ligand or signaling. Further, the GPR124 phenotype indicates significant regional heterogeneity in embryonic CNS requirements for angiogenic signals, which could operate in the context of telencephalic angiogenic gradients, as have been proposed (16). The spatial restriction of GPR124−/− angiogenesis deficits is strongly paralleled by a VEGF-independent activity mediating GPR124- and Cdc42-dependent migration to forebrain but not hindbrain conditioned medium. In principle, this activity could represent ligand or render competence to respond to ligand. However, this by no means excludes GPR124 interactions with non-soluble factors such as ECM (17) or transmembrane proteins, or distinct mechanisms for non-angiogenic receptor functions. The progressive nature of Tie2-GPR124 vascular malformations indicates a continued sensitivity of brain endothelium to GPR124 during adulthood, with the adult temporal window possibly reflecting either moderate gain-of-function with receptor overexpression or differential sensitivity to receptor in adulthood.
Wnt/β-catenin (8, 9), VEGF/Nrp1 (18, 19), integrins αv and β8 (20–22), and Id1/Id3 (23), mutations impair CNS angiogenesis, often with knockout glomeruloid malformations resembling the GPR124 phenotype (24). Regional regulation of angiogenesis has been reported in CXCL12/CXCR4 mutants although not in the CNS (25, 26). However, as opposed to GPR124, none of these loci encode CNS-specific endothelial receptors and typically manifest vascular deficits globally or in subsets of non-CNS organs (27–29). Despite marked phenotypic overlap, GPR124 does not appear to exhibit epistasis with β-catenin (fig. S20), and GPR124 may function in a parallel pathway coupling embryonic CNS angiogenesis to developmental Glut1 expression. Similar epistasis studies with Id1 and integrin β8 have also been negative (fig. S20).
The significant CNS enrichment of endothelial GPR124 expression, as well as the GPR124 gain- and loss-of-function CNS angiogenesis phenotypes suggest the potential relevance of this receptor to various CNS-related vascular pathologies. The conditional floxed mouse GPR124 alleles will enable future investigation of angiogenic functions in disorders such as stroke and malignancy, and in potential developmental or adult BBB functions. Certainly, the determination of potential causal relationships between GPR124 and CNS vascular malformations or germinal matrix hemorrhage (30) in human and mouse should be worthwhile. Finally, the current analyses provide insight into the poorly understood functions of the adhesion receptor class of GPCRs (12), and show how control of angiogenesis can be managed in unique ways by specific organ systems, in this case with insight into angiogenesis in the central nervous system and development of the blood-brain barrier. Similar signaling pathways may be deployed among vascular beds unique to other organ systems.
Supplementary Material
Acknowledgements
We are indebted to Richard Daneman, Ben Barres, Cecile Chartier and the Kuo laboratory for comments, Jenny Yuan and Jessica Hampton for animal husbandry and genotyping, and Andrew Connolly and Hannes Vogel for neuropathology. FK was supported by an AHA Postdoctoral Fellowship and MRM by AHA Predoctoral Fellowship AHA09PRE2110036, and the Stanford MSTP Program. This work was supported by NIH R01NS27713 (WLY) and P01NS444155C (HS), UK MRC G0501711 (MF); NIH 1DP2 OD006477, 1R21 NS058600, and National Academies Keck Futures Initiative CS10 grants (SCH); and NIH 1R01NS052830, 1R01NS064517, 1R01HL074267, Brain Tumor Society, Goldhirsh Foundation, and the Stanford Center for Children’s Brain Tumors grants (CJK).
References
- 1.Carmeliet P. Nature. 2005 Dec 15;438:932. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 2.Aird WC. Circ Res. 2007 Feb 2;100:174. doi: 10.1161/01.RES.0000255690.03436.ae. [DOI] [PubMed] [Google Scholar]
- 3.Ballabh P, Braun A, Nedergaard M. Neurobiol Dis. 2004 Jun;16:1. doi: 10.1016/j.nbd.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Pardridge WM. Methods Mol Med. 2003;89:385. doi: 10.1385/1-59259-419-0:385. [DOI] [PubMed] [Google Scholar]
- 5.Shen Q, et al. Science. 2004 May 28;304:1338. [Google Scholar]
- 6.Hogan KA, Ambler CA, Chapman DL, Bautch VL. Development. 2004 Apr;131:1503. doi: 10.1242/dev.01039. [DOI] [PubMed] [Google Scholar]
- 7.Herken R, Gotz W, Wattjes KH. J Anat. 1989 Jun;164:85. [PMC free article] [PubMed] [Google Scholar]
- 8.Stenman JM, et al. Science. 2008 Nov 21;322:1247. doi: 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- 9.Daneman R, et al. Proc Natl Acad Sci U S A. 2009 Jan 13;106:641. doi: 10.1073/pnas.0805165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.St Croix B, et al. Science. 2000 Aug 18;289:1197. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 11.Carson-Walter EB, et al. Cancer Res. 2001 Sep 15;61:6649. [PubMed] [Google Scholar]
- 12.Fredriksson R, Gloriam DE, Hoglund PJ, Lagerstrom MC, Schioth HB. Biochem Biophys Res Commun. 2003 Feb 14;301:725. doi: 10.1016/s0006-291x(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 13.Claxton S, et al. Genesis. 2008 Feb;46:74. doi: 10.1002/dvg.20367. [DOI] [PubMed] [Google Scholar]
- 14.Schlaeger TM, et al. Proc Natl Acad Sci U S A. 1997 Apr 1;94:3058. doi: 10.1073/pnas.94.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamloo A, Ma N, Poo MM, Sohn LL, Heilshorn SC. Lab Chip. 2008 Aug;8:1292. doi: 10.1039/b719788h. [DOI] [PubMed] [Google Scholar]
- 16.Vasudevan A, Long JE, Crandall JE, Rubenstein JL, Bhide PG. Nat Neurosci. 2008 Apr;11:429. doi: 10.1038/nn2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallon M, Essler M. J Biol Chem. 2006 Nov 10;281:34179. doi: 10.1074/jbc.M605291200. [DOI] [PubMed] [Google Scholar]
- 18.Gerhardt H, et al. Dev Dyn. 2004 Nov;231:503. doi: 10.1002/dvdy.20148. [DOI] [PubMed] [Google Scholar]
- 19.Raab S, et al. Thromb Haemost. 2004 Mar;91:595. doi: 10.1160/TH03-09-0582. [DOI] [PubMed] [Google Scholar]
- 20.McCarty JH, et al. Development. 2005 Jan;132:165. doi: 10.1242/dev.01551. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J, et al. Development. 2002 Jun;129:2891. doi: 10.1242/dev.129.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader BL, Rayburn H, Crowley D, Hynes RO. Cell. 1998 Nov 13;95:507. doi: 10.1016/s0092-8674(00)81618-9. [DOI] [PubMed] [Google Scholar]
- 23.Lyden D, et al. Nature. 1999 Oct 14;401:670. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 24.Mancuso MR, Kuhnert F, Kuo CJ. Lymphat Res Biol. 2008;6:173. doi: 10.1089/lrb.2008.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ara T, Tokoyoda K, Okamoto R, Koni PA, Nagasawa T. Blood. 2005 Apr 15;105:3155. doi: 10.1182/blood-2004-07-2563. [DOI] [PubMed] [Google Scholar]
- 26.Siekmann AF, Standley C, Fogarty KE, Wolfe SA, Lawson ND. Genes Dev. 2009 Oct 1;23:2272. doi: 10.1101/gad.1813509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmeliet P, et al. Nature. 1996 Apr 4;380:435. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 28.Ferrara N, et al. Nature. 1996 Apr 4;380:439. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 29.Cattelino A, et al. J Cell Biol. 2003 Sep 15;162:1111. doi: 10.1083/jcb.200212157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballabh P. Pediatr Res. 2010 Jan;67:1. doi: 10.1203/PDR.0b013e3181c1b176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.