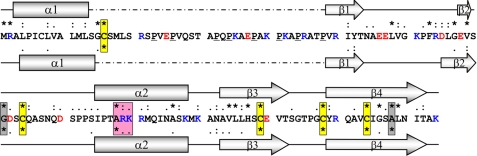
FIGURE 1.
Sequence alignment and secondary structure prediction for E. coli RcsF. The amino acid sequence of E. coli RcsF (P69411) is shown (positively charged amino acids are shown in blue, negatively charged amino acids in red, and prolines are underlined). The top row of symbols represents conservation within Enterica sequences, and the bottom row represents total conservation between all annotated RcsF sequences. The secondary structure elements predicted by the PsiPred server for E. coli RcsF (P69411) and the most distant annotated RcsF sequence from Aeromonas hydrophilia (A0KMN2) are presented above and below the E. coli sequence, respectively. The boxes emphasize conserved residues (yellow, cysteines; red, positive charged cluster; gray, others).