Abstract
Glycosaminoglycans (GAGs) expressed ubiquitously on the cell surface are known to interact with a variety of ligands to mediate different cellular processes. However, their role in the internalization of cationic gene delivery vectors such as liposomes, polymers, and peptides is still ambiguous and seems to be controlled by multiple factors. In this report, taking peptides as model systems, we show that peptide chemistry is one of the key factors that determine the dependence on cell surface glycosaminoglycans for cellular internalization and gene delivery. Arginine peptides and their complexes with plasmid DNA show efficient uptake and functional gene transfer independent of the cell surface GAGs. On the other hand, lysine peptides and complexes primarily enter through a GAG-dependent pathway. The peptide-DNA complexes also show differential interaction with soluble GAGs. In the presence of exogenous GAGs under certain conditions, arginine peptide-DNA complexes show increased transfection efficiency that is not observed with lysine. This is attributed to a change in the complex nature that ensures better protection of the compacted DNA in the case of arginine complexes, whereas the lysine complexes get destabilized under these conditions. The presence of a GAG coating also ensures better cell association of arginine complexes, resulting in increased uptake. Our results indicate that the role of both the cell surface and exogenous glycosaminoglycans in gene delivery is controlled by the nature of the peptide and its complex with DNA.
Keywords: Atomic Force Microscopy, Chondroitin Sulfate, Gene Therapy, Glycosaminoglycan, Heparan Sulfate, Peptides
Introduction
Proteoglycans are biologically important molecules that are ubiquitously expressed on the cell surfaces as well as the extracellular and intracellular spaces and are involved in a variety of functions including tissue organization, morphogenesis, cellular communication, adhesion, migration, growth, and signaling to name few (1). They are made up of a protein core linked to linear polysaccharide chains called glycosaminoglycans (GAGs).3 These glycosaminoglycan chains are composed of different repeating disaccharide units that are sulfated at various positions. Based on the nature of the repeating units and the degree of sulfation, heparin, heparan sulfate, chondroitin sulfates, keratan sulfates, and hyaluronan are the different types of GAGs well known in the literature. Sulfated proteoglycans on the cell membranes are the major contributors to the negative charge on the cell surface, making it available for electrostatic interactions. These proteoglycans bind many ligands like growth factors and chemokines as well as large proteins. They have also been implicated in the internalization of foreign molecules and organisms through endocytotic pathways. Many viruses including adeno-associated virus (2), herpes simplex virus, human papilloma virus, and human immunodeficiency virus (3) initially bind to cell surface heparan sulfate proteoglycans to mediate their cellular entry.
Non-viral gene delivery vectors like lipoplexes (lipid-DNA complexes) and polyplexes (polymer-DNA complexes), which are usually cationic in nature, can also interact with both cell surface and extracellular GAGs through electrostatic interactions. The cell surface GAGs have been thought to act as the primary receptor for the attachment of the cationic complexes followed by endocytosis of the complexes (for review, see Ref. 4). Early reports showed that poly-l-lysine-DNA complexes as well as different cationic liposome-DNA complexes require cell surface proteoglycans (specifically, heparan sulfate proteoglycans) for delivery of DNA both in vitro and in vivo (5, 6). However, many subsequent reports have shown that their function in cellular entry of non-viral vector complexes may be dispensable, and strong binding of cationic complexes to either cell surface or extracellular GAGs may actually prevent their uptake. It was demonstrated that cellular uptake of cationic lipoplexes occurred at similar levels in cell lines with cell surface GAGs as well as in GAG-deficient cell lines (7), which implies that any role of GAGs in controlling transfection efficiency can at best come into play at a later step of the transfection process. It has also been suggested that cell surface proteoglycans protect cells from the cytotoxic effects of cationic lipids, and hence, proteoglycan-deficient cell lines give lowered transfection at high lipid to DNA ratios (7). In contrast, another study showed that cationic polymers such as poly-l-lysine and polyethyleneimine, and lipids such as N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl sulfate (DOTAP) and 1,2-dioleyl-3-phosphatidylethanolamine (DOPE) show strong binding to the cell surface GAGs but decreased transfection efficiency in their presence due to decreased cellular uptake (8). The exact role of the cell surface proteoglycans in lipoplex or polyplex delivery is, thus, elusive, and different experimental conditions as well as different chemical and structural characteristics of the cationic carriers might be responsible for the differences observed in the data.
Exogenous addition of negatively charged GAGs in vitro during transfection has also been studied in most of these systems and has usually been found to decrease the gene transfer efficiency (5, 9). It is thought that depending on the chemistry of the carrier, the nature of the complex, and the charge density of GAGs, the decrease in transfection may be due to one or all of the following reasons. (i) Sulfated GAGs cause premature extracellular release of DNA from its complexes with cationic polymers (10, 11) or cationic lipids (12). (ii) Soluble GAGs could compete with cell surface proteoglycans for binding to the complex, thereby decreasing cellular uptake (13). (iii) Binding of GAGs may change the endocytotic uptake route of the complex. (iv) GAGs may cause altered intracellular distribution of the complexes, significantly affecting the gene expression (9).
The strong and universal translocation of arginine-rich cell-penetrating peptides in multiple cell types has also triggered speculation on the involvement of some common molecules like cell surface GAGs in the process of cellular entry, although it is still debatable whether membrane translocation and endocytotic uptake are both involved (14). Cell-penetrating peptides like naturally occurring protein transduction domains, e.g. the HIV-TAT peptide (13), Antennapedia peptide (15), penetratin, and even synthetic homoarginines of certain compositions (16, 17), are likely to involve cell surface GAGs in their cellular entry. It has also been seen that when these peptides are used for cargo delivery, the requirement of cell surface GAGs differs. Cellular uptake of HIV-1 TAT peptide conjugated to cargo was first surmised to occur in a manner dependent on the presence of heparan sulfate proteoglycans, which was confirmed by impaired uptake on enzymatic or genetic removal of the cell surface GAGs (13, 18). However, uptake of free TAT peptide was suggested to involve either different receptors or pathways because its internalization was not completely inhibited in cells lacking surface heparan sulfate (19). The involvement of cell surface proteoglycans on the cellular entry of the peptide and its complexes with cargo can be affected by many factors such as net charge of the complex (18) as well as the structure and distribution of positive charges on the peptides (17), and hence, it is possible that the bare peptide and the complex behave in different ways. More recently, it was proposed that transduction mediated by TAT can occur in glycan-deficient cell lines and the cell surface glycans bind TAT and only affect the efficiency of transduction (20). In addition, although arginine-rich peptides have high affinity to sulfated GAGs, especially heparan sulfate, and heparin (21) and could co-internalize heparan sulfate, mediating nuclear delivery (18), soluble sulfated GAGs inhibited cargo delivery by arginine-rich peptides (13, 15). All these evidences suggest that the role of GAGs in controlling cellular entry of cationic peptides with and without cargo needs further elucidation.
Under this backdrop, we have explored the role of both cell surface and soluble GAGs during gene delivery using model peptide carriers, arginine, and lysine homopeptides (each 16-mer in length). Peptides containing these two amino acids either alone or in different proportions (e.g. TAT peptide) and often with simple chemical modifications like attachment of other additional amino-acids, polyethylene glycol, functional moieties like targeting ligands, and fusogenic peptides, etc. are the most commonly studied peptide based gene delivery agents (22, 23). We show that the 16-mer arginine and lysine homopeptides differ in their requirement of cell surface GAGs as well as in their interaction with soluble GAGs during gene delivery. This is reflected in the alterations of complex morphology, pattern of cellular binding, internalization, and eventual gene delivery efficiency between the two peptide systems. Our results highlight the importance of peptide chemistry as a key determinant for requirement of GAGs in gene delivery.
EXPERIMENTAL PROCEDURES
Materials
The peptides used in this study were 16-mer of l-lysine (K16) and l-arginine (R16). The unlabeled and fluorescein isothiocyanate (FITC)-labeled peptides (>95% purity) were custom-synthesized by GL Biochem (Shanghai) Ltd. The plasmids pEGFP-C1, 4.7 kb (Clontech) and pMIR-REPORTTM Luciferase, 6.47 kb (Ambion), were amplified in Escherichia coli DH5-α and purified using GenElute HP Endotoxin-Free Plasmid MaxiPrep kit (Sigma). Heparin sodium salt (from porcine intestinal mucosa), heparan sulfate sodium salt (fast moving fraction from porcine intestinal mucosa), chondroitin sulfate A sodium salt (from bovine trachea), chondroitin 6-sulfate (C6S) sodium salt (from shark cartilage), chondroitinase ABC, heparinase III, and DNase I were purchased from Sigma. The luciferase assay kit was from Promega. Label IT® Tracker Fluorescein kit for labeling plasmid DNA was purchased from Mirus Bio Corp. All other fine chemicals and cell culture media were from Sigma.
Cell Culture
Parental Chinese hamster ovary cells (CHO-K1) were obtained from the National Centre for Cell Science Cell Repository, India. The glycosaminoglycan mutant cell line pgsA-745 was obtained from the American Type Culture Collection. Both cell lines were maintained in Ham's F12K medium supplemented with 10% (v/v) fetal bovine serum (Invitrogen) in a humidified 5% CO2, 37 °C incubator.
Preparation of Peptide-Plasmid DNA Complex
Peptide-DNA complexes (polyplexes) were prepared at different charge ratios expressed as peptide nitrogen per nucleic acid phosphate (N/P) or as Z(+/−). The DNA stock was diluted to a concentration of 20–40 ng/μl and added dropwise to an equal volume of the appropriate peptide dilution while vortexing. The polyplexes were incubated for 30 min at room temperature before performing any experiment.
Transfection and Luciferase Gene Expression Assay
Cells were seeded 24 h before transfection in 24-well plates at a density of 50,000 cells per well. Polyplexes were prepared at Z(+/−) of 10 with a final DNA concentration of 20 ng/μl (pMIR-ReportTM Luciferase) and incubated for 1 h at room temperature. Indicated amounts of GAGs, expressed as GAG:peptide (w/w), were added to the complexes after 30 min of incubation and kept for further 30 min. 100 μl of polyplex (2 μg of DNA/well) was added to cells (70% confluency) in serum-free media (Opti-MEM, Invitrogen). After 5 h of incubation at 37 °C, the medium was aspirated, and cells were washed with phosphate-buffered saline (PBS, pH 7.4) and supplemented with 500 μl of complete growth medium. After 24 h of transfection, cells were washed with PBS and lysed with 100 μl of cell culture lysis buffer (1× CCLR, Promega). Luciferase expression was measured in 50 μl of cell lysate supernatant using the luciferase assay substrate (Promega). Light emission was measured by integration over 10 s in Orion microplate luminometer (Berthold Detection System, Germany). Luciferase activity was normalized with total protein content of the cells, estimated using BCA protein assay (Pierce).
Treatment of Cells with Chlorate and GAG Lyases
CHO-K1 cells were seeded in 24-well plates and treated with chlorate or GAG lyases after 24 h. For desulfation of GAGs, cells were incubated in media supplemented with sodium chlorate (NaClO3, 75 mm) for a further 24 h. For removal of cell-associated GAGs, cells were treated with either chondroitinase ABC (250 milliunits) or heparinase III (2 mIU) in 300 μl of digestion buffer (PBS containing 0.1% BSA, 0.2% gelatin, and 0.1% glucose) (13) for 1 h at 37 °C. Cells were washed extensively with PBS and serum-free media before transfection was carried out as detailed above.
Labeling of Plasmid DNA with FITC
Plasmid DNA (pEGFP-C1) was labeled with FITC using the Label IT® Tracker Fluorescein kit (Mirus Bio Corp.) at a 0.75:1 (v:w) ratio, i.e. 0.75 μl of labeling reagent/μg of DNA according to manufacturer's protocol.
Flow Cytometry Analysis
Cells were grown for 24 h in 24-well plates and fluorescently labeled polyplexes (DNA labeled with FITC) at Z(+/−) of 10, or FITC-labeled peptides (5 μm) were added to cells in serum-free media as detailed in the transfection protocol. After 4 h of incubation at 37 °C or 1 h at 4 °C, cells were washed twice with ice-cold PBS containing 1 mg/ml heparin to remove the extracellular-bound polyplex and with 0.4% trypan blue in PBS to quench the extracellular fluorescence wherever required. Cells were collected by trypsinization (100 μl of 0.25% trypsin) and resuspended in 500 μl of PBS and placed on ice. Flow cytometry measurements were carried out on Guava® EasyCyteTM System (Guava Technologies) using CytoSoftTM software. 10,000 live cells were used for each analysis.
For analyzing the binding of peptides or polyplexes to cell membranes, cells were washed with serum-free medium and placed on ice for 20 min before the addition of polyplexes. Cells were incubated with the peptides or polyplexes for 1 h on ice. Medium was removed, and cells were washed twice with ice-cold PBS, scraped, and resuspended in 500 μl PBS for flow cytometry analysis as mentioned above.
Stability of Polyplexes to GAGs Determined by Agarose Gel Electrophoresis
The polyplexes formed at charge ratio Z(+/−) of 10 (20 μl containing 200 ng DNA) were treated with increasing amounts of GAGs (heparin, heparan sulfate, chondroitin sulfate A, and chondroitin 6-sulfate) and incubated for 30 min before loading on 1% agarose gel. Electrophoresis was carried out at 100 V in Tris acetate-EDTA buffer for 30 min. The amount of the DNA released from the polyplexes was compared with that of the native uncomplexed DNA.
Stability of Polyplexes to GAGs Determined by Ethidium Bromide (EtBr) Intercalation Assay
Polyplexes were prepared at Z(+/−) of 10 and incubated for 30 min at room temperature. GAGs, in increasing amounts, were added to black 96-well plates (Nunc) followed by the addition of 20 μl of polyplex and 10 μl of EtBr (4.22 ng/μl) and incubation for 5 min at room temperature in the dark. Fluorescence intensity was measured in a DTX 880 Multimode detector (Beckman Coulter) using 535 SL EXP 1 excitation and 595 SL EMP 1 emission filters. The fluorescence of DNA with EtBr was taken as the maximum, i.e. 100%, and the relative percentage increase in fluorescence signal was calculated at increasing concentration of GAGs.
DNase I Assay
DNase I was reconstituted in buffer containing 50 mm Tris-HCl, pH 7.5, 1 mm CaCl2, 10 mm MgCl2, and 50% glycerol. The polyplexes at charge ratio 10 were incubated with different concentrations of GAGs for 30 min at room temperature. The polyplexes with GAGs were then treated with DNase I (1 units) for 30 min at 37 °C in reaction buffer consisting of 10 mm Tris-HCl, pH 7.5, 0.1 mm CaCl2, and 2.5 mm MgCl2. DNase I was inactivated by heating at 75 °C for 10 min. To release the protected DNA from the polyplex, heparin (5 μg) was added to the above reaction mixture and was further incubated for 30 min at room temperature. The mixture was then analyzed on 1% agarose gel. The integrity and amount of DNA protected was compared with DNA released from those polyplexes (after the addition of heparin) that were not subjected to DNase treatment.
Atomic Force Microscopy
The polyplexes at Z(+/−) 10 with or without GAGs were imaged by depositing 2 μl of the polyplex solution on freshly cleaved mica and drying it in air. Appropriate controls of GAGs were also similarly imaged. Imaging was carried out with 5500 scanning probe microscope (Agilent Technologies, Inc.) using PicoView software. Images were obtained in tapping mode in air with 225-μm-long silicon cantilevers (Agilent Technologies) that have a resonance frequency of around 75 kHz and a force constant of 2.8 newtons/m. Scan speed used was 1 line/s. Minimum image processing (first order flattening and brightness contrast) was employed. Image analysis was performed using PicoImage software.
Confocal Microscopy
Cells were seeded at a density of 1.2 × 105 in 35-mm μ-dishes (ibidi, Germany) and incubated for 24 h. FITC-labeled peptides (10 μm) were added to the cells in serum-free media and incubated at 4 °C for 1 h. Cells were washed three times with ice-cold PBS(+) containing heparin (1 mg/ml). Imaging was done on an inverted LSM510 META laser scanning microscope (Carl Zeiss) using a Plan-Apochromat 63 × 1.4 N.A. lens and the 488-nm line of an argon laser.
RESULTS
Arginine and Lysine Homopeptides Show Nearly Similar Transfection Efficiency in the Presence and Absence of Cell Surface Glycosaminoglycans
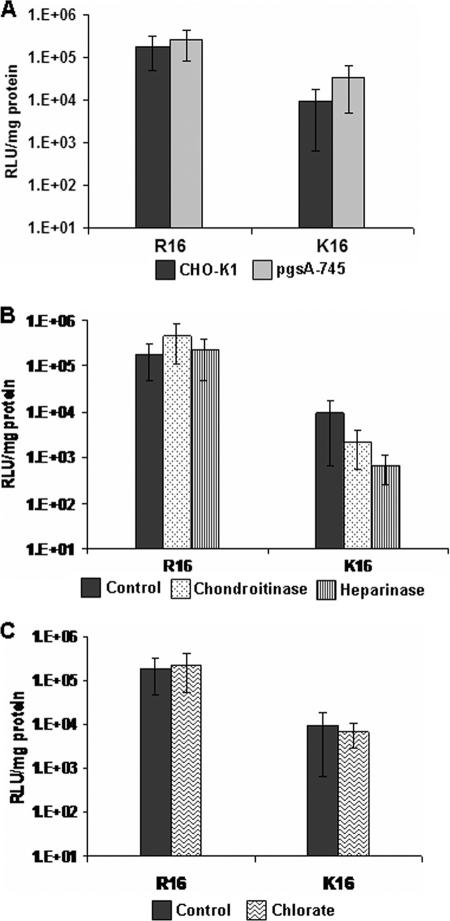
To explore the role of cell surface GAGs in DNA delivery efficiency, we first studied the efficiency of transfection of plasmid DNA with R16 and K16 peptides in wild type CHO-K1 and the glycosaminoglycan-deficient pgsA-745 (mutant with xylosyl transferase deficiency (24) producing <10% cell surface proteoglycans (8)) cell lines at a charge ratio Z(+/−) = 10. We have seen that at this charge ratio, maximum transfection efficiency was achieved with minimal toxicity in the case of both the peptides and thus allows for the best comparison between the two (data not shown). The transfection efficiency of R16 in both the cell lines is higher than that of K16 (which has been rationalized by us in an earlier study.4 The control data in CHO-K1 in some of the experiments are adapted from Mann et al.4 The transfection efficiency changes only marginally for both the peptides in the two cell lines as shown in Fig. 1A. Although the difference was more evident in the case of K16, it is difficult to conclusively establish the importance of GAGs because of the overall low transfection efficiency of lysine peptides. In addition, CHO-K1 cells were pretreated with the enzymes heparinase III and chondroitinase ABC for selective enzymatic removal of cell surface heparan sulfate (HS) and chondroitin sulfate (CS) to study the transfection efficiency of R16 and K16 peptides under selective presence of one type of GAG on the cell surface. The results are shown in Fig. 1B. In the case of R16, there was a mild increase in transfection efficiency by up to 2 times after chondroitinase treatment. However, transfection efficiency of K16 was slightly more affected by the absence of cell surface GAGs; in particular there was a considerable decrease in the transfection efficiency after enzymatic removal of HS. This might imply that the cell surface GAGs are required for transfection in the case of lysine peptides. Altogether, these results seem to suggest that GAGs associated with the cell surface proteoglycans do not have any strong inhibitory effect on the transfection efficiency, particularly in the case of R16. We also carried out transfection in the wild type cell line after treatment with sodium chlorate that prevents sulfation of the GAGs. As shown in Fig. 1C, transfection occurred efficiently in the absence of sulfation, but it was not evident whether the cell surface GAGs are dispensable or whether the K16 polyplexes utilize attachment sites other than the sulfate groups of GAGs.
FIGURE 1.
Transfection efficiency of R16 and K16 polyplexes in GAG-deficient conditions. A, wild type CHO-K1 and its glycan-deficient mutant pgsA-745 were treated with R16 and K16 polyplexes. Transfection efficiency was measured from luciferase activity at 24 h after 5 h of incubation of the cells with the polyplexes. B, CHO-K1 cells were pretreated with either chondroitinase ABC (250 milliunits) or heparinase III (2 mIU) for 1 h at 37 °C before transfection was carried out. C, for desulfation of GAGs, CHO-K1 cells were treated with sodium chlorate (75 mm) for 24 h before transfection. Values are given as mean ± S.D. RLU, relative light units.
Cellular Uptake of Arginine and Lysine Peptides and Polyplexes Show Distinctly Different Dependence on Cell Surface Glycosaminoglycans
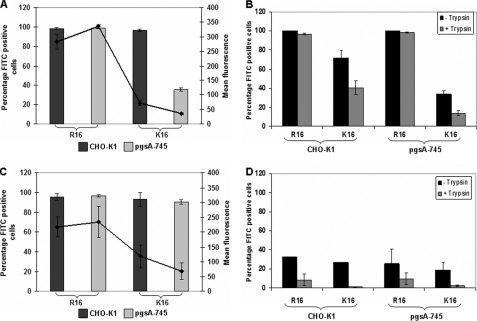
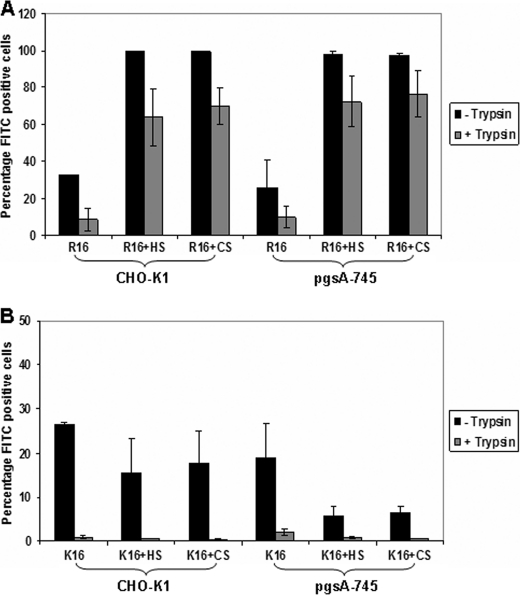
To check whether GAGs affect the cellular entry process, we investigated the uptake of both the free peptide and the polyplex in the two cell lines using flow cytometry. Both free peptides enter into 99% cells in the wild type cell line. Uptake of free R16 peptide (labeled with FITC) is independent of cell surface GAGs as seen by a similar percentage of fluorescence-positive cells as well as their mean intensity in the two cell lines (Fig. 2A). In contrast, the presence of cell surface proteoglycans seems to be favorable for the uptake of K16, as indicated by a decrease of about 60% in the number of fluorescence-positive cells as well as a drop in the fluorescence intensity in the GAG-deficient cell line. This shows that the free K16 peptide might be primarily utilizing a GAG-dependent route for cellular entry, although entry is not altogether abolished in the absence of cell surface GAGs. We have also studied the cell association and internalization of the free peptide at low temperature, where endocytotic uptake is likely to be absent (Fig. 2B). Cell association refers to both the membrane-bound and the internalized peptides, as cells that were dissociated non-enzymatically were used for the analysis. To determine only the internalized fraction, cells were washed with heparin (1 mg/ml in PBS) to remove the extracellular-bound peptides (26) and additionally with trypan blue in PBS to quench any extracellular fluorescence and treated with trypsin to remove the surface-bound peptides (27). In the case of R16, the peptide was able to internalize in nearly the entire cell population in both the cell lines, showing that R16 adopts a non-endocytotic route for direct entry into the cells that is independent of cell surface GAGs. On the other hand, in the case of K16, although cell association was seen in about 70% of the cells in the case of CHO-K1, the peptide internalized in only about 40% of the CHO-K1 cells, which could be indicative of both a GAG-dependent endocytotic entry and a less predominant non-endocytotic route, where only the latter is operative at low temperatures. In the GAG-deficient cells (Fig. 2B), the association and internalization is much lower, indicating that cell surface GAGs act as primary attachment sites for lysine peptides. This was further confirmed by studying the localization of the peptides at 4 °C in the wild type and mutant cell lines by confocal microscopy. R16 showed cytoplasmic staining in most of the cells in both cell types, whereas K16 was internalized in a small number of wild type cells and very few GAG-deficient cells (supplemental Fig. 1).
FIGURE 2.
Cellular association and uptake of R16 and K16 in wild type and GAG-deficient CHO cells. A, FITC-labeled R16 and K16 peptides (5 μm) were added to CHO-K1 and pgsA-745 cells. After 4 h of incubation at 37 °C, cells were washed with ice-cold PBS containing 1 mg/ml heparin and with 0.4% trypan blue in PBS. Cells were collected by trypsinization and analyzed by flow cytometry. B, FITC-labeled R16 and K16 peptides (5 μm) were added to CHO-K1 and pgsA-745 cells. After 1 h of incubation at 4 °C, cells were washed with ice-cold PBS and removed by non-enzymatic cell dissociation for analysis of cell association by flow cytometry (black bars). To determine internalization, cells were washed with ice-cold PBS containing 1 mg/ml heparin and with 0.4% trypan blue in PBS after incubation with peptides. Cells were collected by trypsinization and analyzed by flow cytometry (gray bars). C, R16 and K16 polyplexes containing FITC-labeled plasmid DNA were added to the cells and treated as described in A. D, R16 and K16 polyplexes containing FITC-labeled plasmid DNA were added to the cells and treated as described in B. Bars represent the percentage of fluorescence positive cells, and lines represent the mean fluorescence intensity in arbitrary units for each sample. Values are given as the mean ± S.D.
We next studied the uptake of the polyplexes at charge ratio 10.0 prepared with FITC-labeled plasmid DNA and unlabeled peptide. Uptake of R16 polyplexes exhibited similar levels in both the cell lines as shown in Fig. 2C as in the case of the free peptide. However, at 4 °C, R16 polyplexes showed very low cell association and even lower internalization in both the cell lines (Fig. 2D), indicating that the entry is primarily endocytotic, unlike that in the case of the bare peptide. In the case of K16 polyplexes, however, uptake at 37 °C showed variations in the two cell lines; although the number of fluorescence-positive cells is similar, there is a 40% drop in mean fluorescence intensity in the fluorescence-positive cells in the GAG-deficient cell line (Fig. 2C). This was further confirmed by checking the complex uptake when the CHO-K1 cells were treated with heparinase and chondroitinase for selectively removing the cell surface GAGs. K16 complexes showed a drop in uptake in both the treated conditions, unlike the R16 complexes (data not shown). The cellular association of K16 polyplexes, determined at 4 °C, is comparable with that of R16 complexes; however, negligible internalization was observed in either cell line (Fig. 2D), indicating that the entry process is entirely endocytotic in case of K16 complexes.
Thus, the results above indicate that in the case of R16, the cell surface GAGs do not have any effect on the uptake of the bare peptide or its complex with DNA (although they seem to utilize different modes of entry), nor do they affect the transfection efficiency. In case of K16, however, the peptide and complex uptake occur either partially or completely through the endocytotic route, respectively, and require GAGs on the cell surface. However, transfection by K16 can still occur in the GAG-deficient cell line.
The Addition of Exogenous Glycosaminoglycans Enhances Transfection Efficiency under Several Conditions in the Case of Arginine Peptides but Not in Lysine Peptides
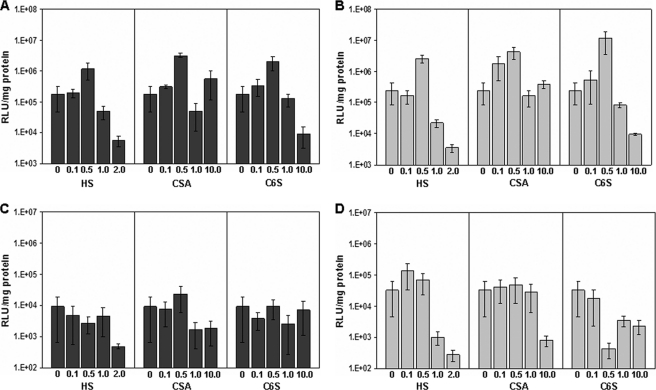
We next wanted to study whether this difference in the behavior of arginine and lysine polyplexes is also seen during their cellular delivery in the presence of exogenously added GAGs. We carried out transfection in both the cell lines with R16 and K16 polyplexes where different concentrations of HS, CS-A, and chondroitin-6-sulfate have been added to the polyplexes as described under “Experimental Procedures.” The transfection efficiencies in the CHO-K1 cell line for both the peptides are shown in Fig. 3, A and C, and those carried out in the pgsA-745 cell line are shown in Fig. 3, B and D. In the case of R16, when the GAG was added in low concentration (i.e. 1 and 5 μg per 100 μl of polyplex at Z = 10.0, corresponding to w/w ratios of GAG:peptide of 0.1:1 and 0.5:1 respectively), there was an increase in the transfection efficiency, albeit to different levels in both the cell lines. The addition of HS at a GAG:peptide ratio of 0.5:1 led to a 6× increase in transfection, whereas the addition of the same concentration of CS resulted in enhancement of transfection by more than 10× in CHO-K1 cells (Fig. 3A). The increase was even more striking in the GAG-deficient cell line (Fig. 3B; 10–20× increase at these concentrations). The addition of still higher concentrations of any of the GAGs led to a considerable decrease in transfection efficiency. Such a substantial increase in transfection efficiency at low concentrations of the GAGs was, however, not seen in the case of K16. In the wild type CHO-K1 cells (Fig. 3C), there was a decrease in transfection efficiency on the addition of GAGs across the entire concentration range studied, with the exception of a marginal increase 2.5 times over the base value on the addition of CS-A at a GAG:peptide ratio of 0.5:1.When the GAG-deficient cell line was used (Fig. 3D), there was a mild increase (2–3×) in transfection efficiency with HS at very low amounts, whereas the transfection efficiency remained nearly constant or decreaseed with different concentrations of both the types of CS.
FIGURE 3.
Effect of exogenous glycosaminoglycans on the transfection efficiency of R16 and K16. Increasing amounts of GAGs (HS, CSA, and C6S; expressed as GAG:peptide w/w ratio) were added to R16 (A and B) and K16 (C and D) polyplexes. CHO-K1 (dark gray bars) and pgsA-745 (light gray bars) cells were treated with the polyplexes for 5 h at 37 °C, and luciferase gene expression was measured after 24 h. Values are given as the mean ± S.D. RLU, relative light units.
This difference in the transfection efficiency by the arginine and lysine peptides in the presence of exogenous GAGs could happen because of differences in the interaction of the two types of polyplexes with the free GAGs or in their cellular uptake and intracellular processing. We first examined whether the interaction with free GAGs affects the nature and stability of the polyplexes in vitro on the addition of GAGs and further looked for possible differences in the uptake and internalization efficiencies in the presence of free GAGs.
Arginine Polyplexes Are More Stable Than Lysine Polyplexes in the Presence of Different Glycosaminoglycans in Vitro
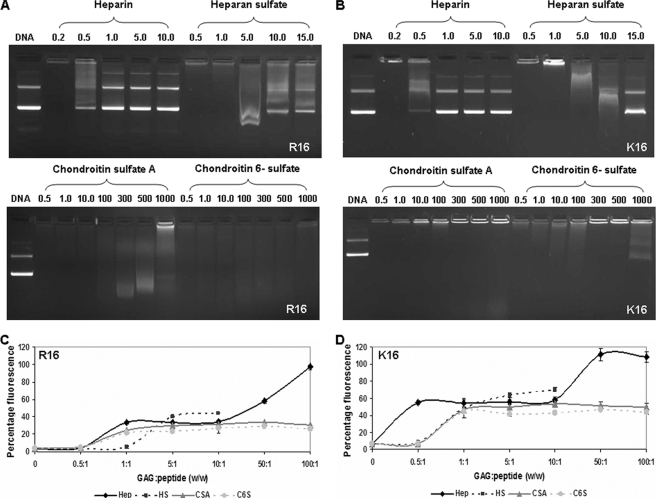
Stability of the polyplexes on anionic perturbation with different GAGs was first analyzed using agarose gel electrophoresis (Fig. 4, A and B). It was seen that both the polyplexes of R16 and K16 show near complete stability toward CS with very mild DNA release at very high concentrations (at GAG:peptide w/w of 300:1 and above). In the presence of the more negatively charged HS, both polyplexes showed DNA release beyond a ratio of 5:1, whereas heparin with the highest number of negative charges tends to release DNA from the polyplexes at much lower amounts. However, it is significant to note that for both R16 and K16, the concentration of GAG where either a slight or significant increase in transfection efficiency has been obtained (GAG:peptide ratio of 0.1:1 and 0.5:1), both polyplexes seemed to be completely condensed from the agarose gel electrophoresis study.
FIGURE 4.
Stability of R16 and K16 polyplexes to anionic challenge by GAGs. R16 and K16 polyplexes were treated with increasing amounts of different GAGs (heparin, HS, CSA, and C6S) expressed as GAG:peptide w/w ratios. Stability was checked as a measure of the DNA released by agarose gel electrophoresis (the amount of DNA released was compared with uncomplexed plasmid DNA in lane 1 in A and B) and an ethidium bromide intercalation assay (the amount of DNA released was measured by the increase in the fluorescence by intercalation of EtBr; C and D). Fluorescence of free plasmid complexed to EtBr is taken as 100%. Values are plotted as percentage of maximum ± S.D.
We further examined the stability of the polyplexes by analyzing the DNA release patterns using the EtBr intercalation assay. When the polyplexes were subjected to an anionic challenge, they destabilized (loosened structures or completely released DNA), and the fluorescence recovery on re-intercalation of the EtBr in DNA was a measure of the relative amount of DNA released. It was seen from Fig. 4, C and D, that the addition of CS over the entire concentration range studied resulted in a maximum fluorescence recovery of 30% in the case of R16 polyplexes and up to 50% in the case of K16 polyplexes. Heparin showed a near complete release in both the cases. In the case of HS, both the polyplexes are destabilized, but once again the percentage of recovery was more in the case of K16 (60%) as compared with R16, where the recovery was about 40%. This indicates that although the DNA release efficiency depends on the negative charge density on the GAG (heparin > HS > CS), there are subtle differences detected only in the more sensitive EtBr assay between R16 and K16 polyplexes toward each agent. Clearly, K16 polyplexes are more easily destabilized in the presence of all the GAGs as compared with R16 polyplexes.
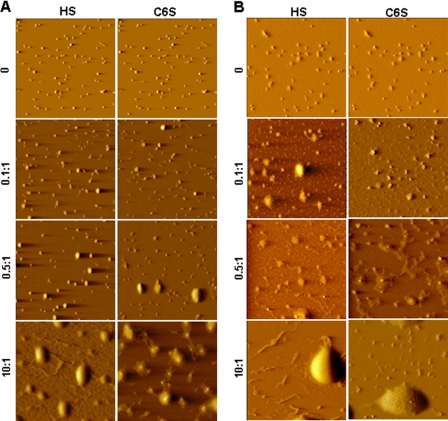
We also used atomic force microscopy to study the changes in the morphology of the complexes formed with R16 and K16 in the presence of increasing concentrations of HS and CS. R16 polyplexes (Fig. 5A) did not show any change in morphology at low amounts (GAG:peptide ratio of 0.1:1) of either HS or CS. At slightly higher amounts (0.5:1), “loosening” of the complexes was seen in the form of flower-shaped structures in a small population. In contrast, there was destabilization as well as DNA release for K16 polyplexes at all the concentrations of added HS or CS (Fig. 5B). At very high amounts of the GAG (10:1, where the transfection efficiency drops for both R16 and K16), both the polyplexes were disassembled to a large extent as expected.
FIGURE 5.
Morphologies of R16 and K16 polyplexes treated with GAGs as seen by atomic force microscopy. Polyplexes of R16 (A) and K16 (B) were treated with increasing amounts of HS and C6S expressed as GAG:peptide w/w ratio. 2 μl of the resulting complex was deposited on mica, air-dried, and imaged in air with an atomic force microscope.
These experiments conclusively establish that any change in the polyplex nature in the presence of GAGs depends on the nature of the peptide used to form the polyplex, the GAG concentration, and the nature of the GAG. In the case of R16, the complex morphology was not significantly affected by very low GAG concentration, whereas there was DNA release from the K16 polyplexes at the same concentrations. Because the increase in transfection efficiency of R16 was also seen with lower concentrations of GAG, it is possible that the presence of GAG offers better stability to the polyplex. Therefore, we tested the extent of protection of the complexed DNA against nucleases using DNase I protection assay. As shown in Fig. 6, in the case of R16 polyplexes, the presence of a lower amount (GAG:peptide w/w of 0.5:1) of GAG offered more protection to the complexed DNA than the case where no GAG was present. It is only at a higher amount (1:1) that the GAGs tend to destabilize the complexes to some extent, making the DNA more accessible to nuclease attack. But for K16 polyplexes, GAGs do not confer any protection to the DNA, as even small quantities of the anionic agent destabilize the polyplex.
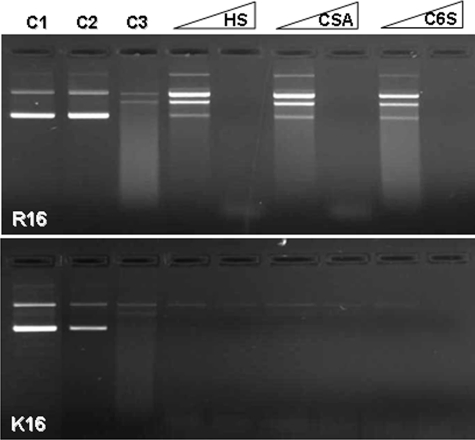
FIGURE 6.
Effect of GAGs on the DNA protection ability of R16 and K16 polyplexes against DNase I. R16 and K16 polyplexes were treated with GAGs (HS, CSA, and C6S) at 0.5:1 and 1:1 GAG:peptide w/w. Complexes were then subjected to DNase 1 (1 unit) treatment. The amount of DNA protected was then released by heparin and checked by agarose gel electrophoresis. Controls: lane C1, free plasmid; lane C2, polyplex treated with heparin; lane C3, polyplex treated with DNase I and heparin.
The results from the in vitro experiments thus clearly show that the R16 polyplexes are more stable in the presence of GAGs, and the DNA remained better protected at the concentrations of GAGs where an increase in the transfection efficiency was seen. The K16 polyplexes on the other hand were easily destabilized and amenable to nuclease attack in presence of the GAGs.
Arginine and Lysine Polyplexes Show Differences in Cellular Association and Uptake in the Presence of Exogenous Glycosaminoglycans
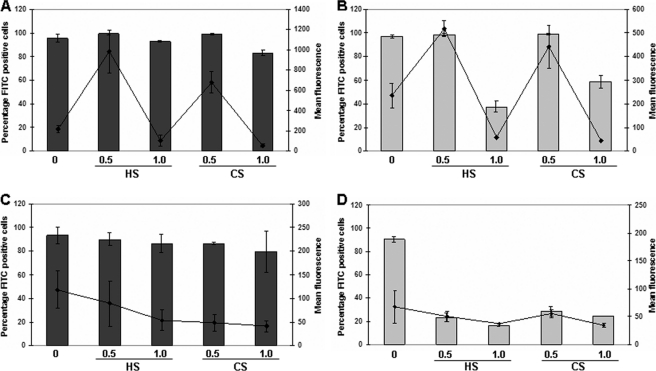
We next investigated whether these differences in the polyplex characteristics in the presence of GAGs also affect their association to the cell membranes of the wild type and the mutant cell lines in the presence of HS and CS. Cells were incubated with polyplexes for 1 h at 4 °C, and the extent of cell association and internalization of the polyplexes was determined using flow cytometry as described under “Experimental Procedures.” The addition of GAGs (at GAG:peptide w/w ratio of 0.5:1) increased the cellular association in both the CHO-K1 and mutant cell lines for the R16 polyplexes (Fig. 7A). There was a strong increase in the number of fluorescence-positive cells (>95% positive cells with GAGs compared with 30% without GAGs). However, in the case of K16 (Fig. 7B), the presence of exogenous GAGs reduced the association of the polyplexes considerably in both the cell lines. We also determined the extent of internalized polyplexes in the presence and absence of GAGs (at 0.5:1) in both the cell lines after trypsin treatment, which removes the surface-bound polyplexes. Although R16 polyplexes showed internalization in less than 10% cells in the absence of GAGs, the addition of GAGs increased the percentage of fluorescence-positive cells to more than 60% that in both cell lines. The mutant cell line showed slightly higher internalization (Fig. 7A). But for K16 polyplexes, the presence of exogenous GAGs did not promote internalization (Fig. 7B). These results indicate a preferential role of the exogenous GAGs in the association and internalization of the R16 polyplexes, whereas such an effect is absent for the K16 polyplexes.
FIGURE 7.
Effect of exogenous GAGs on the cellular association and internalization efficiency at 4 °C of R16 and K16 polyplexes. R16 (A) and K16 (B) polyplexes containing FITC-labeled plasmid DNA were treated with HS and CS at 0.5:1 GAG:peptide w/w. Complexes were added to CHO-K1 and pgsA-745 cells and incubated for 1 h at 4 °C. To measure the cellular association efficiency (black bars), cells were washed with ice-cold PBS, removed non-enzymatically, and resuspended in PBS for flow cytometry analysis. To compare the internalized polyplexes (gray bars), cells were washed with ice-cold PBS containing 1 mg/ml heparin and then with 0.4% trypan blue in PBS. Cells were collected by trypsinization and analyzed by flow cytometry. Values are given as the mean ± S.D.
The cellular uptake of the arginine and lysine polyplexes in the presence of GAGs was also determined after incubation for 4 h at 37 °C. The uptake of the polyplexes under similar conditions in the absence of GAGs (as shown in Fig. 2C) was quite high in R16 polyplexes. As shown in Fig. 8, the addition of 5 μg (ratio of 0.5:1) of HS or CS in R16 polyplexes led to a 4.5× and 3× increase in the fluorescence intensity, respectively, in CHO-K1 cells (Fig. 8A), whereas the increase was nearly twice in the mutant cell line (Fig. 8B). Higher amounts of GAGs caused a significant decrease in the number of cells that took up the polyplexes, particularly in the mutant cell line, corresponding with earlier observations. In the case of K16, there was a drop in the percentage of positive cells and their mean fluorescence intensity on the addition of GAGs at all concentrations (Fig. 8, C and D), indicating that the presence of exogenous GAGs inhibits the entry of K16 polyplexes due to destabilization of the polyplexes. Thus, arginine- and lysine-based peptide-DNA complexes behave differently in the presence of exogenous GAGs, which reflects in their cellular entry and transfection efficiencies.
FIGURE 8.
Effect of exogenous GAGs on the cellular uptake of R16 and K16 polyplexes. R16 (A and B) and K16 (C and D) polyplexes containing FITC-labeled plasmid DNA were treated with HS and CS at 0.5:1 and 1:1 GAG:peptide w/w. Complexes were added to CHO-K1 (dark gray) and pgsA-745 (light gray) cells. After 4 h of incubation at 37 °C, cells were washed with ice-cold PBS containing 1 mg/ml heparin and with 0.4% trypan blue in PBS. Cells were collected by trypsinization and analyzed by flow cytometry. Values for controls are same as in Fig. 2C. Bars represent the percentage of fluorescence-positive cells, and lines represent the mean fluorescence intensity in arbitrary units on the secondary axis. Values are given as the mean ± S.D.
DISCUSSION
Cell surface glycosaminoglycans, by virtue of their negative charge density, are known to act as initial attachment sites or receptors for the internalization of a variety of ligands. However, although these molecules seem to be important in the entry of cell penetrating peptides like TAT, Antp, and some oligoarginines, a general consensus on their exact role is lacking. Multiple factors seem to affect the interaction between the positively charged peptides and the cell surface GAGs and eventually their internalization mechanism. The peptide sequence, charge distribution and the extracellular peptide concentration are the major influential factors (28) along with the composition of the cell surface or the presence of other receptors. It has also been observed that the peptide conjugated to a cargo, either covalently or electrostatically, behave differently or may have alternative internalization mechanisms as compared with the free peptide (19). With respect to the peptide complexes with cargo molecules like DNA, the role of cell surface GAGs is ambiguous and seems to affect various stages of cellular entry like complex stability and cell surface binding as well as intracellular processing. Moreover, free exogenous GAGs may hinder the gene delivery efficiency by peptide-DNA complexes, as has already been reported for lipoplexes and polyplexes (5, 9).
In this study, our aim was to explore the role of cell surface and exogenous GAGs on the DNA delivery efficiency of arginine and lysine homopeptides of similar length, with the main focus on the role of peptide chemistry in this process. These two amino acids are the most common residues in peptide-based carriers for gene delivery and were thus obvious choices for a comparative study. Our results indicate that arginine- and lysine-based peptides differ in their requirement of cell surface GAGs for both cellular entry as well as DNA delivery. In the case of R16, the free peptide and the polyplex appear to adopt different routes of internalization. The arginine peptide enters entirely by non-endocytotic pathway as observed by near complete internalization at low temperature. On the other hand, R16 polyplexes go through an endocytotic mechanism. However, it is interesting to note that in both cases, there is very little difference in the uptake or the transfection efficiency between the wild type and the glycan-deficient cell lines or under the conditions of enzymatic removal of cell surface GAGs (as seen from Figs. 1 and 2), indicating that the peptide and polyplex probably utilize different entry paths, which are both non-GAG-dependent. In contrast, both the bare lysine homopeptide and its complex are taken up into the cells primarily through an endocytotic route of entry involving cell surface GAGs. There is a reduction in the uptake of both the K16 peptide and polyplex in the mutant cell line and a decrease in the transfection efficiency on enzymatic removal of cell surface GAGs (both HS and CS). This implicates GAGs as primary attachment sites for the polyplexes, which is confirmed by poor cell association and internalization at low temperature in the mutant cell line. However, the transfection efficiency in the mutant cell line is not hugely altered. This apparent discrepancy could arise from a step beyond the entry process, i.e. possible routing through a different intracellular processing pathway in the mutant cell line as compared with the wild type, leading to efficient gene expression. Altogether, these results indicate that a glycan-dependent pathway of entry seems to be the predominant mode for the lysine peptide.
The cellular entry of arginine-rich peptides has been proposed to occur by multiple pathways, including direct translocation across the membrane and endocytosis (29, 30). Both the pathways were operational in the proteoglycan-deficient cell line as well, although at higher peptide concentration, showing that the cell surface proteoglycans are dispensable for cellular entry of arginine homopeptides (31). The polar, positively charged arginine oligomers can recruit negatively charged membrane components such as fatty acids to transiently produce a less polar ion pair complex that can partition into the lipid bilayer and thus translocate across the plasma membrane, driven by the membrane potential (32). This is due to the more effective bidentate hydrogen bonding possible for guanidino groups in arginine versus the mondentate hydrogen bonding for ammonium groups in lysine. Therefore, lysine peptides form weaker interactions with membrane components, and direct translocation of lysine is not efficient (32, 33). It is possible that such a mechanism of entry is operative here as well in case of the arginine peptides and polyplexes, whereas the lysine counterpart employs a GAG-dependent endocytotic route.
We have also checked the transfection efficiency of the two peptides in the presence of exogenous soluble GAGs to explore whether there are differences in the interaction of arginine and lysine peptides with GAGs. Surprisingly, we observed that in the presence of exogenous HS and CS, the transfection efficiency of R16 increases by an order of magnitude or more under certain concentrations of GAGs (Fig. 3), which is contrary to some of the earlier reports. The increase is seen at relatively low GAG:peptide w/w ratios of 0.1:1 and 0.5:1 and is independent of cell surface GAGs. At relatively higher amounts of GAGs, there is a decrease in transfection, as has been observed earlier, possibly by destabilization of the complexes (13, 15). On the other hand, in the case of K16 polyplexes, the addition of exogenous GAGs does not in general cause any major increase in the transfection efficiency and shows considerable drop at most of the GAG concentrations used.
To explain this, we analyzed the stability of these polyplexes on anionic challenge. In vitro analysis of the stability of the polyplexes in the presence of soluble GAGs by agarose gel electrophoresis and ethidium bromide exclusion assay shows that both types of polyplexes are destabilized only at very high amounts of HS or CS (significantly higher than the conditions under which increased transfection is seen). The extent of destabilization is higher in the case of K16 polyplexes. However, atomic force microscopy reveals that there are actually subtle changes in the polyplex morphology in the two cases in the presence of GAGs. R16 complexes show altered morphology in the presence of GAGs but do not release DNA, whereas the K16 polyplexes clearly release DNA at all concentrations of anionic challenge despite the fact that arginines are known to bind more strongly to GAGs than lysines (34). DNase I protection assay also confirms that low amounts of GAGs confer stability to the R16 polyplexes making the compacted DNA unavailable for nuclease degradation, as compared with the native polyplexes without GAGs. However, such protection is not offered in the case of K16 polyplexes. Thus, clearly the K16 complexes, by virtue of their loosened nature, are easily disassembled in presence of exogenous GAGs, which could be the major reason for their lowered transfection in such conditions.
The differences in the interaction of arginine and lysine polyplexes with soluble GAGs can be ascribed to differences in the nature of the complexes formed by these peptides with DNA. We have earlier observed in a separate study that arginine- and lysine-based peptides have different DNA delivery efficiency due to differences in their DNA compaction and release mechanism. At a charge ratio 10, where maximum transfection efficiency (with least toxicity) is seen with both the peptides, the polyplexes show small size and uniform size distribution in the case of R16, whereas the particles are loosely packed in case of K16 (this is also shown in Fig. 5 under conditions where GAGs are absent). We have attributed this to the fact that K16 shows multiple modes of complexation (both monomolecular and multimolecular pathways are operative) as compared with R16 (where only multimolecular pathways are seen). The release of DNA from the polyplexes on anionic challenge also corroborates this, as K16 is more easily destabilized in the presence of anionic agents.4 It has also been shown in the literature that arginine polypeptide binds with higher affinity to different glycosaminoglycans like heparan sulfate and chondroitin sulfate than lysine, as the guanidino group of arginine can form electrostatic interaction as well as hydrogen bonds with the sulfate groups in GAGs when compared with the ammonium cation of lysine (25, 35). For a similar reason, arginine polypeptide also binds with higher affinity to a negatively charged polymer like DNA. Thus, the stronger affinity for GAGs allows the more compact R16 complexes to accommodate low amounts of GAGs on the surface, leading to increase in size but minimal destabilization (Fig. 5A) along with increased transfection efficiency, whereas the loose packaging of K16 polyplexes is easily disturbed in presence of GAGs. It needs to be noted here that in terms of charge, 5 μg of HS has ∼5 times lesser negative charges than the positive charges on R16 peptides in a complex with DNA. Thus, if we assume that the GAGs attach to the complex surface, the polyplex still remains positively charged in the presence of the GAGs and possibly has a protective effect.
We further dissected at which step the GAG protected R16 polyplexes score over the K16 polyplexes during cellular interaction. We observed that in the presence of GAGs at low concentrations (0.5:1 GAG:peptide) the R16 polyplex-GAG complex was associated with more than 95% of cells (at 4 °C) in the case of both the wild type and the glycan-deficient mutant cells as compared with only 30% cells when no GAG is present. This shows that the R16 polyplexes, in the presence of a GAG coating on them, show more cell association, but the binding sites possibly do not involve cell surface GAGs, as the effect is seen in both the wild type and the mutant cell line. Polyplexes were also internalized more efficiently in the presence of GAG coating both at 4 and 37 °C. Because internalization at 4 °C is usually indicative of a non-endocytotic route of entry, it is possible that multiple uptake routes are utilized by the GAG-coated R16 complexes. On the other hand, K16 polyplexes show decreased cell association and uptake in the presence of exogenous GAGs, which is even more prominent in the mutant cell line. Once again, this confirms the need for cell surface GAGs for the internalization of K16 polyplexes. But whether the cell surface proteoglycans are directly involved in the internalization of polyplexes by endocytosis or they act as primary receptors to present the complexes to specific endocytotic receptors is not clear and needs to be elucidated further.
In summary, in this manuscript we have shown that arginine and lysine peptides have distinct behaviors in the presence of glycosaminoglycans. 16-Mer arginine homopeptides and their complexes with DNA can enter glycan-deficient cells as efficiently as the wild type cells, showing that cell surface GAGs are dispensable for their entry. But 16-mer lysine homopeptides and complexes prefer a GAG-dependent pathway for cellular entry. In the presence of relatively lower amounts of soluble GAGs, the arginine polyplexes show better stability and give an increase in the transfection efficiency in both cell types, unlike the lysine polyplexes. The chemical nature of the peptide and their different mechanisms of DNA compaction and release can thus cause differences in their behavior in the presence of both cell surface and soluble GAGs. The interaction between R16 and different GAGs does not seem to show specificity, as the consequential effects of R16 polyplex binding to GAGs remains similar for both HS and CS. The specificity might arise from differences in peptide length and structural characteristics as it has been demonstrated that the structures of arginine-rich peptides, specifically the distribution of positive charges, determine their dependence and specificity for heparan sulfate proteoglycan (17). In addition to stabilization of the complexes and their higher membrane binding affinity and uptake in the presence of GAGs, arginine polyplexes might also be adopting a different route of uptake or intracellular processing which delivers functional DNA more efficiently. This aspect is currently under investigation. We are also trying to explore whether conjugating GAG to peptide-DNA complexes will be useful in targeting specific cell types or whether this modulation will enhance transfection efficiency in a GAG-rich environment.
Supplementary Material
This work was supported by the Council of Scientific and Industrial Research, Government of India (Project NWP35).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
A. Mann, G. Thakur, V. Shukla, A. K. Singh, R. Khanduri, R. Naik, Y. Jiang, N. Kalra, B. S. Dwarakanath, U. Langel, and M. Ganguli, submitted for publication.
- GAG
- glycosaminoglycan
- HS
- heparan sulfate
- CS
- chondroitin sulfate
- C6S
- chondroitin 6-sulfate.
REFERENCES
- 1. Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 2. Summerford C., Samulski R. J. (1998) J. Virol. 72, 1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen Y., Götte M., Liu J., Park P. W. (2008) Mol. Cells 26, 415–426 [PubMed] [Google Scholar]
- 4. Poon G. M., Gariépy J. (2007) Biochem. Soc. Trans. 35, 788–793 [DOI] [PubMed] [Google Scholar]
- 5. Mislick K. A., Baldeschwieler J. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12349–12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mounkes L. C., Zhong W., Cipres-Palacin G., Heath T. D., Debs R. J. (1998) J. Biol. Chem. 273, 26164–26170 [DOI] [PubMed] [Google Scholar]
- 7. Belting M., Petersson P. (1999) Biochem. J. 342, 281–286 [PMC free article] [PubMed] [Google Scholar]
- 8. Ruponen M., Honkakoski P., Tammi M., Urtti A. (2004) J. Gene Med. 6, 405–414 [DOI] [PubMed] [Google Scholar]
- 9. Ruponen M., Rönkkö S., Honkakoski P., Pelkonen J., Tammi M., Urtti A. (2001) J. Biol. Chem. 276, 33875–33880 [DOI] [PubMed] [Google Scholar]
- 10. Ruponen M., Ylä-Herttuala S., Urtti A. (1999) Biochim. Biophys. Acta 1415, 331–341 [DOI] [PubMed] [Google Scholar]
- 11. Burke R. S., Pun S. (2008) Bioconjugate Chem. 19, 693–704 [DOI] [PubMed] [Google Scholar]
- 12. Zelphati O., Szoka F. C., Jr. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11493–11498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tyagi M., Rusnati M., Presta M., Giacca M. (2001) J. Biol. Chem. 276, 3254–3261 [DOI] [PubMed] [Google Scholar]
- 14. Futaki S., Nakase I., Tadokoro A., Takeuchi T., Jones A. T. (2007) Biochem. Soc. Trans. 35, 784–787 [DOI] [PubMed] [Google Scholar]
- 15. Console S., Marty C., García-Echeverría C., Schwendener R., Ballmer-Hofer K. (2003) J. Biol. Chem. 278, 35109–35114 [DOI] [PubMed] [Google Scholar]
- 16. Fuchs S. M., Raines R. T. (2004) Biochemistry 43, 2438–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakase I., Tadokoro A., Kawabata N., Takeuchi T., Katoh H., Hiramoto K., Negishi M., Nomizu M., Sugiura Y., Futaki S. (2007) Biochemistry 46, 492–501 [DOI] [PubMed] [Google Scholar]
- 18. Sandgren S., Cheng F., Belting M. (2002) J. Biol. Chem. 277, 38877–38883 [DOI] [PubMed] [Google Scholar]
- 19. Richard J. P., Melikov K., Brooks H., Prevot P., Lebleu B., Chernomordik L. V. (2005) J. Biol. Chem. 280, 15300–15306 [DOI] [PubMed] [Google Scholar]
- 20. Gump J. M., June R. K., Dowdy S. F. (2010) J. Biol. Chem. 285, 1500–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rusnati M., Coltrini D., Oreste P., Zoppetti G., Albini A., Noonan D., d'Adda di Fagagna F., Giacca M., Presta M. (1997) J. Biol. Chem. 272, 11313–11320 [DOI] [PubMed] [Google Scholar]
- 22. Martin M. E., Rice K. G. (2007) AAPS J. 9, E18–E29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mann A., Thakur G., Shukla V., Ganguli M. (2008) Drug Discov. Today 13, 152–160 [DOI] [PubMed] [Google Scholar]
- 24. Esko J. D., Stewart T. E., Taylor W. H. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3197–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verrecchio A., Germann M. W., Schick B. P., Kung B., Twardowski T., San Antonio J. D. (2000) J. Biol. Chem. 275, 7701–7707 [DOI] [PubMed] [Google Scholar]
- 26. Lundberg M., Wikström S., Johansson M. (2003) Mol. Ther. 8, 143–150 [DOI] [PubMed] [Google Scholar]
- 27. Richard J. P., Melikov K., Vives E., Ramos C., Verbeure B., Gait M. J., Chernomordik L. V., Lebleu B. (2003) J. Biol. Chem. 278, 585–590 [DOI] [PubMed] [Google Scholar]
- 28. Jiao C. Y., Delaroche D., Burlina F., Alves I. D., Chassaing G., Sagan S. (2009) J. Biol. Chem. 284, 33957–33965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duchardt F., Fotin-Mleczek M., Schwarz H., Fischer R., Brock R. (2007) Traffic 8, 848–866 [DOI] [PubMed] [Google Scholar]
- 30. Nakase I., Takeuchi T., Tanaka G., Futaki S. (2008) Adv. Drug Delivery Rev. 60, 598–607 [DOI] [PubMed] [Google Scholar]
- 31. Kosuge M., Takeuchi T., Nakase I., Jones A. T., Futaki S. (2008) Bioconjugate Chem. 19, 656–664 [DOI] [PubMed] [Google Scholar]
- 32. Rothbard J. B., Jessop T. C., Wender P. A. (2005) Adv. Drug Delivery Rev. 57, 495–504 [DOI] [PubMed] [Google Scholar]
- 33. Mitchell D. J., Kim D. T., Steinman L., Fathman C. G., Rothbard J. B. (2000) J. Pept. Res. 56, 318–325 [DOI] [PubMed] [Google Scholar]
- 34. Hileman R. E., Fromm J. R., Weiler J. M., Linhardt R. J. (1998) Bioessays 20, 156–167 [DOI] [PubMed] [Google Scholar]
- 35. Fromm J. R., Hileman R. E., Caldwell E. E., Weiler J. M., Linhardt R. J. (1995) Arch. Biochem. Biophys. 323, 279–287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.