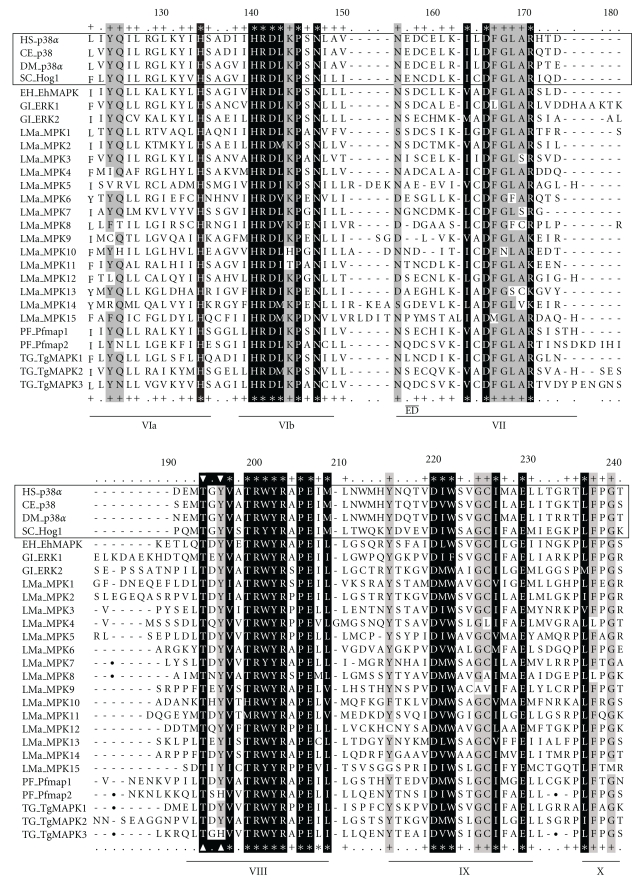
Figure 1.
ClustalW alignment of representative MAPKs of diverse evolutionary origin, with each of the 11 subdomains indicated (Roman numerals). Conserved acidic residues within the ED site (subdomain VII) and common docking (CD) domain, which immediately follows subdomain XI, have been underlined. The first four sequences represent p38 MAPKs of metazoan and yeast origin and are boxed as reference sequences to which other protozoan MAPKs can be compared. Invariant MAPK residues (within allowed substitution groups) are highlighted in black and denoted by an asterisk. Highly conserved residues (>80% conservation) are highlighted in grey and denoted by a plus sign. In the absence of grey shading, plus signs indicate residues conserved in the majority of aligned sequences. Allowed substitution groups include acidic/amide (DE, DN, EQ), aliphatic (LIVM), aromatic (FYW), basic (KR), and hydroxyl/polar residues (STG). The positions of insertion sequences removed prior to ClustalW alignment are indicated by filled circles. White triangles denote the position of the TX[XY] phosphorylation lip. Two letter abbreviations precede the name of each MAPK sequence, indicating the genus and species of origin for each MAPK. CE: Caenorhabditis elegans; DM: Drosophila melanogaster; EH: Entamoeba histolytica; GI: Giardia intestinalis; HS: Homo sapiens; LMa: Leishmania major; PF: Plasmodium falciparum; SC: Saccharomyces cerevisiae; TG: Toxoplasma gondii. Accession numbers of all aligned sequences are listed in Tables 1 and 2.