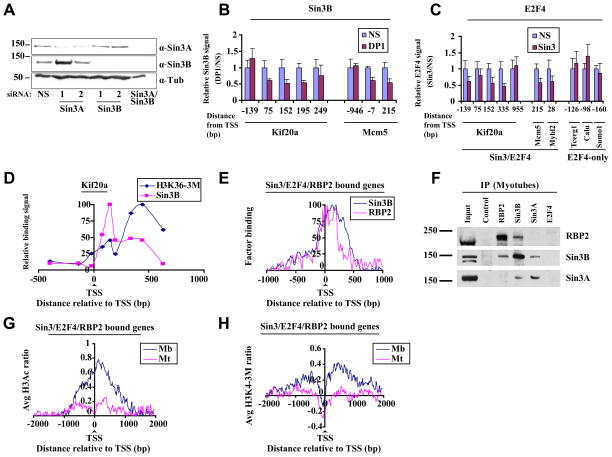
Figure 3. Sin3/E2F4-RBP2 function as a complex on target genes.
(A) Representative western blot indicates knock-down of Sin3A and Sin3B after transfection of myotubes with indicated siRNAs. Myotubes were transfected 48 h after induction of differentiation and were isolated 72h after transfection. (B) Histogram depicting Sin3B binding assessed by qChIP of selected genes in differentiated myotubes transfected with a non-specific control (NS) and DP1 siRNAs. Relative binding is expressed as a ratio of signals obtained in specific vs. NS siRNA transfected cells. The average of three independent experiments is shown. Error bars represent standard deviation. (C) As in B except that cells were transfected with control and Sin3A/Sin3B (Sin3) siRNAs and E2F4 binding was measured. (D) H3K36 tri-methylation levels and Sin3B binding were examined on the Kif20a gene using qChIP. Enrichment was calculated as percentage of input and normalized to maximum binding levels. The results of three independent experiments are shown. (E) Binding profiles for RBP2 and Sin3B binding to genes bound by Sin3, E2F4, and RBP2, as described in Fig. 2D. Only genes bound by all three factors were examined. (F) Endogenous Sin3B and RBP2 interact in vivo. Sin3 and RBP2 were immunoprecipitated using whole cell extracts of differentiated C2C12 cells (myotubes) with the respective antibodies and probed for either RBP2 or Sin3. (G) Genome-wide histone H3 acetylation (H3Ac) and (H) histone H3 lysine 4 tri-methylation (H3K4-3M) profiles of genes bound by Sin3, E2F4, and RBP2 in myoblasts and differentiated myotubes were obtained using tiling arrays. Genes were aligned relative to the TSS, and average histone H3Ac or H3K4-3M values per probe were calculated. The results of two independent experiments are shown.