Summary
The Dam1 complex attaches the kinetochore to spindle microtubules and is a processivity factor in vitro [1, 2]. In Saccharomyces cerevisiae, which has point centromeres that attach to a single microtubule, deletion of any Dam1 complex member results in chromosome segregation failures and cell death [3–5]. In Schizosaccharomyces pombe, which has epigenetically-defined regional centromeres that each attach to 3–5 kinetochore microtubules, Dam1 complex homologs are not essential [6]. To ask why the complex is essential in some organisms and not in others, we used Candida albicans, a multimorphic yeast with regional centromeres that attach to a single microtubule [7]. Interestingly, the Dam1 complex was essential in C. albicans, suggesting that the number of microtubules per centromere is critical for its requirement. Importantly, by increasing CENP-A expression levels, more kinetochore proteins and microtubules were recruited to the centromeres, which remained fully functional. Furthermore, Dam1 complex members became less crucial for growth in cells with extra kinetochore proteins and microtubules. Thus, the requirement for the Dam1 complex is not due to the DNA-specific nature of point centromeres. Rather, the Dam1 complex is less critical when chromosomes have multiple kinetochore complexes and microtubules per centromere, implying that it functions as a processivity factor in vivo as well as in vitro.
Results and Discussion
Dam1 complex homologs in C. albicans are essential for viability
The Dam1 (DASH) complex is comprised of 10 proteins and has important roles in maintaining the connection between kinetochore proteins and spindle microtubules [8]. Although the Dam1 complex is broadly conserved in fungi [9], its contribution to chromosome segregation differs in different organisms. In the budding yeast S. cerevisiae, deletion of any Dam1 complex member results in chromosome segregation defects and cell death [3–5]. In S. pombe, Dam1 complex members are also kinetochore components, yet they are not essential for viability [6]. Several differences between centromeres and kinetochores in S. cerevisiae and S. pombe could explain the divergent requirement for the Dam1 complex components despite their conservation among fungi. S. cerevisiae has small (~150 bp) point centromeres in which DNA sequence specifies kinetochore position, while S. pombe has larger (40–100 kb) regional centromeres that are determined epigenetically [9]. There are some kinetochore proteins found specifically in organisms with point centromeres (e.g., Ndc10) and others founds specifically in organisms with regional centromeres (e.g., CENP-H/Fta3) [9]. Differences in kinetochore protein composition may affect the requirement for the Dam1 complex. Alternatively, the requirement of the Dam1 complex members for viability may be a matter of one kinetochore-microtubule attachment vs. multiple kinetochore-microtubule attachments per centromere. Kinetochores in S. cerevisiae contain a single nucleosome containing CENP-A, the centromere specific histone H3 isoform, and attach to a single spindle microtubule [10], whereas S. pombe kinetochores are comprised of multiple CENP-A nucleosomes and form attachments to 2–3 spindle microtubules per centromere [7].
To ask why the requirement for the Dam1 complex differs in different fungi, we used the pathogenic yeast C. albicans, which has small (3–10kb), yet regional centromeres, with an average of four CENP-A nucleosomes that attach to a single spindle microtubule per kinetochore [7]. First, we identified putative C. albicans homologs of Dam1 complex members (Figure 1A and Table S1). The C. albicans Dad1 homolog is an intrinsically disordered protein with a structure similar to that of S. cerevisiae Dad1 [11]. By chromatin immunoprecipitation (ChIP), Dad1-GFP and Dam1-GFP bind centromere DNA regions that co-localize with regions bound by CENP-A (Figure 1B). Additionally, Dad1-GFP and Dad2-GFP localized by fluorescence microscopy as discrete spots within DAPI-stained nuclei; the localization pattern of Dad1-GFP and Dad2-GFP was highly similar to that of other kinetochore proteins, such as CENP-A-GFP and Mtw1-GFP (Figure 1C). Together, these results support the conclusion that the genes identified encode Dam1 complex member homologs in C. albicans.
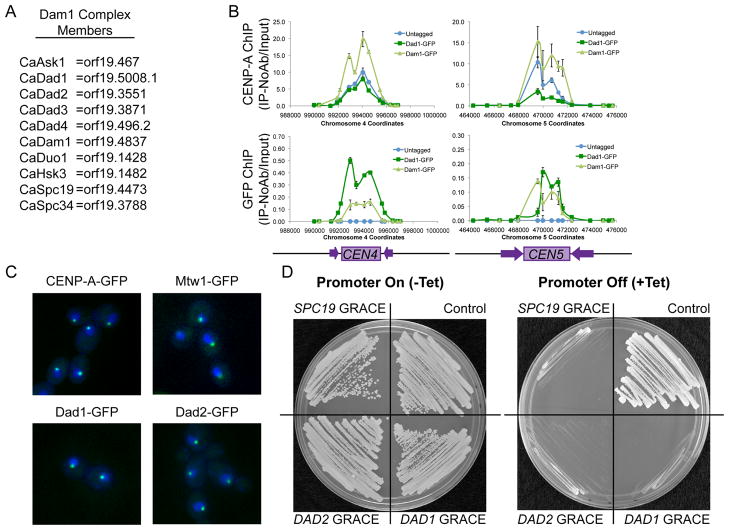
Figure 1. C. albicans Dam1 complex proteins bind centromere DNA regions and are essential in C. albicans.
(A) C. albicans Dam1 complex homologs identified by reciprocal protein-protein BLAST and Pfam family analysis (see also Table 1S). (B) ChIP with anti-CENP-A (upper panel) and anti-GFP antibodies (lower panel) at CEN4 and CEN5 DNA regions in BWP17 (untagged, blue), Dad1-GFP (dark green), and Dam1-GFP (light green) strains shows co-localization of CENP-A and Dam1 complex members on centromeric DNA. Data shown are mean ± SEM of qPCR analysis of each primer set in duplicate and are representative of 3 biological replicates. (C) GFP-tagged CENP-A, Mtw1-GFP, Dad1-GFP and Dad2-GFP (green) show similar localization patterns within DAPI-stained nuclei (blue). Cells were imaged by fluorescence microscopy at 1000x total magnification. (D) GRACE (Gene Replacement and Conditional Expression) DAD1, DAD2, or SPC19 strains and the control strain (SC5314) all exhibited robust growth on SDC-Ura media (left panel), but failed to grow when the Tet-off promoter was repressed by the addition of 50μg/ml tetracycline to SDC-Ura media (right panel).
We next asked if Dam1 complex members are required for viability in C. albicans, an organism with regional centromeres. As C. albicans does not undergo meiosis [12], tetrad analysis cannot be used to determine essentiality. Three independent methods examining four gene products consistently found that Dam1 complex members are required for viability in C. albicans. First, GRACE (Gene Replacement and Conditional Expression) strains [13], in which the only remaining copy of DAD1, DAD2, or SPC19 was conditionally regulated by a Tet-off promoter system, failed to grow when expression was repressed (Figure 1D). Second, when one copy of DAM1 or DAD2 was deleted and the MET3 promoter was used to regulate the remaining copy, the strains did not grow under repressive conditions (media with methionine and cysteine) (Figure S1A). Activation and repression of the promoter in each conditional promoter system was confirmed by quantitative reverse-transcriptase PCR (Figure S1B) and all strains grew as well as control strains under non-repressing conditions (Figure 1D and Figure S1A). Third, we used a UAU single transformation approach [14] to determine that DAD2 and SPC19 are required for viability in C. albicans (Figure S1C-E). Together, these experiments indicate that Dam1 complex members are essential in C. albicans. Thus, the Dam1 complex members are not only required for viability in S. cerevisiae, which has point centromeres, but also in C. albicans, which has regional centromeres.
Extra kinetochore proteins and spindle microtubules localize to centromeres when CENP-A is overexpressed
To test whether the requirement of Dam1 complex members is due to the single kinetochore-microtubule attachment in both S. cerevisiae and C. albicans versus the multiple kinetochore-microtubule attachments per centromere in S. pombe, we needed to develop a method to increase the number of kinetochore-microtubule attachments in C. albicans. This approach is not possible in S. cerevisiae where the number of kinetochore-microtubule attachments is limited by the presence of a single CENP-A nucleosome. CSE4, which encodes the centromeric-specific histone H3 isoform CENP-A in C. albicans [15], is at the top of the kinetochore assembly hierarchy [16, 17], and is closely associated with centromere DNA [18] (Figure 2A). CSE4 expression was conditionally increased by at least 5-fold by placing one copy of the CSE4 gene under control of the PCK1 promoter [19], which is repressed by growth in glucose and induced to high levels by growth in succinate (Figure S2A). Overexpression of CSE4 increased centromeric binding of CENP-A to 293±12% of wild-type levels as measured by ChIP (Figure 2B).
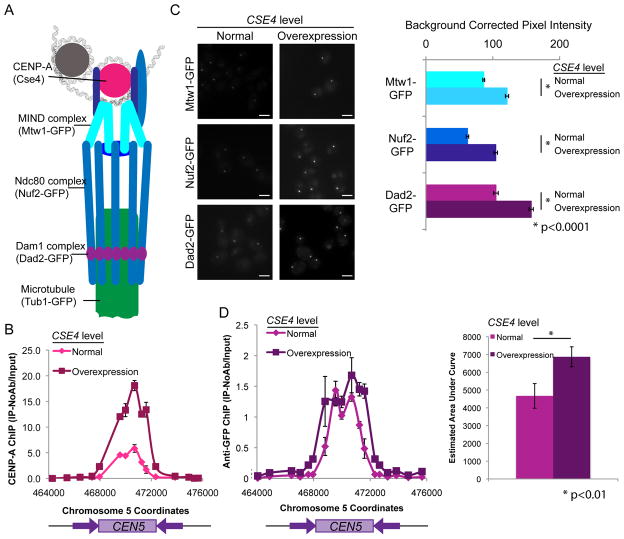
Figure 2. Extra kinetochore proteins localize to centromeres when CENP-A is overexpressed.
(A) Schematic of kinetochore protein organization (adapted from [18]). (B) Anti-CENP-A ChIP analyzed with primers amplifying CEN5 for CENP-A/PCK1p-CENP-A strains grown in repressing conditions (YPA-glucose - normal expression, magenta) and grown in activating conditions (YPA-succinate - overexpression, maroon) for 6 h. Data shown are mean ± SEM of each primer set in duplicate and are representative of 3 biological replicates. (C) CENP-A/PCK1p-CENP-A strains with Mtw1-GFP, Nuf2-GFP or Dad2-GFP were grown in repressing conditions (SDC-glucose - normal expression, left image panels) and grown in activating conditions (SDC-succinate- overexpression, right image panels) for 6 h. Cells were imaged at 1000X total magnification with a GFP filter set. Scale bar = 2μm. GFP fluorescence was quantified by selecting the kinetochore region in each cell and measuring total pixel intensity in the region, corrected for background fluorescence. Data shown are mean ± SEM for at least 100 cells/experiment for 3 biological replicates. Differences between normal expression of CSE4 and overexpression of CSE4 for each strain were found to be statistically significant (<0.0001) using two-tailed unpaired Student’s t-tests (*). (D) PCK1p-CSE4/CSE4 strains with Dad1-GFP were grown in repressing conditions (YPA-glucose - normal expression) and grown in activating conditions (YPA-succinate- overexpression) for 6 h. ChIP with performed with anti-GFP antibodies to assess binding of Dad1-GFP at CEN5 DNA regions. Data shown in the left panel are mean ± SEM of qPCR analysis of each primer set in duplicate and representative of 3 biological replicates. In the right panel, the mean ± SEM of the estimated area under the curve for the CEN5 region for the 3 replicates is shown. Differences between repressing and inducing conditions were found to be statistically significant (p<0.01) using a two-tailed paired Student’s t-test.
Interestingly, CENP-A binding remained localized to the centromere and did not spread to neighboring DNA (Figure 2B). C. albicans centromere regions normally have 4 CENP-A nucleosomes per centromere [7]. The 3-fold increase in CENP-A binding to the centromeric DNA would result in approximately 12 CENP-A nucleosomes per centromere. Interestingly, this is close to the maximum number of nucleosome occupancy sites (~10–12) predicted to be present in C. albicans centromere regions [20]. Consistent with this model, increased association of CENP-A with centromeres results in decreased binding of canonical H3 to the region as measured by ChIP (Figure S2B). When CENP-A was overexpressed in S. cerevisiae, CENP-A remained bound only at the point centromere DNA and was removed from other regions by proteasome-dependent degradation [21]. Increased amounts of CENP-A also appear to remain at least mostly associated with centromeric regions in S. pombe, as CENP-A-GFP overexpression resulted in increased fluorescence signal from kinetochore foci [7]. In contrast to fungi, overexpression of CENP-A in Drosophila [22] and in vertebrate cells [23] can result in CENP-A mislocalization and spurious kinetochore formation on chromosome arms. These differences between species may be due to differences in levels of overexpression; alternatively, fungi may exclude CENP-A from non-centromeric regions more efficiently than metazoans.
Overexpression of CSE4 using a conditional promoter increased the recruitment of the inner kinetochore protein Mtw1 to centromere regions in C. albicans [24]. Thus, we asked whether overexpression of CSE4 would result in increased recruitment of other kinetochore proteins as well. We examined the localization of representative proteins of the MIND complex (Mtw1-GFP), Ndc80 complex (Ndc80-GFP and Nuf2-GFP) and Dam1 complex (Dad1-GFP and Dad2-GFP) to kinetochore foci using quantitative microscopy under conditions of normal and high levels of CSE4 expression (Figure 2C and Figure S2C). The average fluorescence intensity, corrected for background fluorescence, and maximum fluorescence intensity of each GFP-tagged kinetochore protein increased significantly when CSE4 was overexpressed (p<0.01). The average amount of fluorescence intensity that localized to kinetochore foci ranged from 129% to 168% of the average fluorescence intensity with normal CSE4 expression (Figure 2C and Figure S2D). The maximum fluorescence intensity at the kinetochore regions when CSE4 was overexpressed ranged from 140% to 212% of the maximum fluorescence intensity with normal CSE4 expression. ChIP analysis of Mtw1-GFP and Dad1-GFP binding to centromere DNA regions confirmed that kinetochore protein binding to centromeres was enhanced by CSE4 overexpression (Figure 2D and Figure S2E). Dad1-GFP binding at CEN5, as measured by estimating the area under the curve, significantly increased by approximately 1.5 fold (p<0.01), consistent with the increases in fluorescence intensity observed (Figure 2D, right panel). In contrast, control cells without the PCK1 promoter driving CSE4 expression exhibited much smaller differences in fluorescent dot intensity. The background-corrected fluorescence intensity observed for control cells grown in succinate media ranged from 96% to 122% of the intensity observed when cells were grown in glucose (Figure S2F).
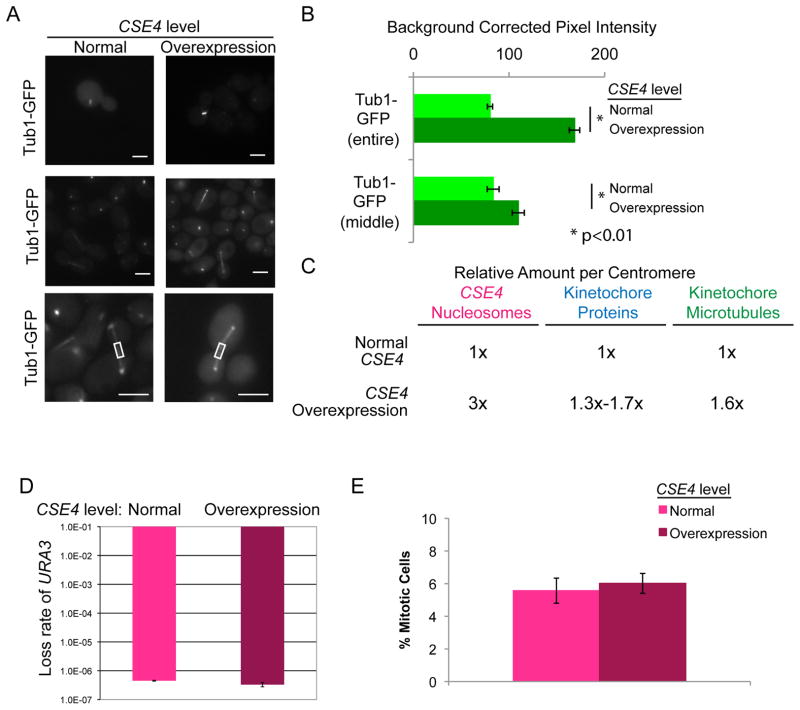
Importantly, we observed increased numbers of microtubules per mitotic spindle when CSE4 was overexpressed (greater than 2-fold higher total Tub1-GFP fluorescence intensity) (Figure 3A and Figure 3B), consistent with extra kinetochore proteins recruiting extra microtubules. During anaphase, the kinetochore-microtubules contract and the fluorescence maximum for the kinetochore-microtubules is found near the spindle pole bodies while the interpolar microtubule fluorescence is concentrated at the center of the spindle [25]. To estimate the increase in interpolar microtubules relative to kinetochore-microtubules, we measured the fluorescence intensity of Tub1-GFP at the center of each anaphase spindle (Figure 3A and Figure 3B, lower panels). When CSE4 was overexpressed, the fluorescence intensity at the center of each spindle increased to 131% of the intensity observed with normal CSE4 expression, compared to 210% of the control intensity when the entire spindle was measured. These results suggest that a significant fraction of the extra spindle microtubules are kinetochore-microtubules, rather than interpolar microtubules. Based on these results, approximately 60% of centromeres have two kinetochore-microtubule attachments when CSE4 is overexpressed in C. albicans (Figure 3C).
Figure 3. Additional spindle microtubules localize to centromeres when CENP-A is overexpressed.
(A) CSE4/PCK1p-CSE4 strains with Tub1-GFP were grown in repressing conditions (SDC-glucose - normal expression, left image panels) and grown in activating conditions (SDC-succinate- overexpression, right image panels) for 6 h. Cells were imaged at 1000X total magnification with a GFP filter set. Upper panels show representative metaphase spindles and middle panels show representative anaphase spindles. Lower panels show increased magnification and example middle regions measured. Scale bar = 2μm. (B) GFP fluorescence was quantified by selecting the mitotic spindle region in each cell (upper bars) or the middle of the mitotic spindle region (lower bars) and measuring total pixel intensity in the region, corrected for background fluorescence. Data shown are mean ± SEM for at least 50 cells/experiment for 4 biological replicates. The difference in mitotic spindle GFP fluorescence between normal expression of CSE4 and overexpression of CSE4 was statistically significant (p<0.0001 for entire spindles, p<0.01 for middle of spindles) using a two-tailed unpaired Student’s t-test (*). (C) Summary of estimated number of kinetochore proteins and kinetochore-microtubule attachments with normal CSE4 expression and overexpression of CSE4. (D) Fluctuation analysis of loss of URA3 in CENP-A/PCK1p-CENP-A strains during growth in repressing conditions (YPA-glucose - normal expression) and growth in activating conditions (YPA-succinate - overexpression). Loss of URA3 was quantified by plating cells on non-selective media (YPA-Glucose) to obtain total numbers of cells and on media containing 5-FOA to select for loss of URA3. Colony counts were used to calculate the rate of FOA/cell division. Results are the mean ± standard deviation of the rates calculated from two experiments, each with 20 cultures per condition. (E) CSE4/PCK1p-CSE4 strains with Tub1-GFP were grown as in (A). The percentage of cells with mitotic spindles was counted. Data shown are mean ± SEM.
Overexpression of CSE4 resulted in increased localization of kinetochore proteins and microtubules to centromere regions; total protein levels of Mtw1-GFP, Nuf2-GFP, Dad2-GFP, and Tub1-GFP remained constant (Figure S3). When measuring the background-corrected pixel intensity of kinetochore proteins and microtubules, we observed that the average background fluorescence in cells grown in succinate was higher than in cells grown in glucose even in the absence of the PCK1-CSE4 construct. To confirm that the increased fluorescence was a difference in background fluorescence as a result of the media and not an increase in protein that was not detected by Western blot, we measured the background fluorescence of an untagged wild-type strain. Background fluorescence was indeed increased in succinate media relative to glucose media (23% increase, p<0.0001) (Figure S3B) indicating that while protein concentrations increased at kinetochore foci, total amounts of kinetochore proteins were not altered.
These results indicate that overexpression of CSE4 in C. albicans increased the recruitment of inner and outer kinetochore proteins to the centromere and resulted in increased numbers of spindle microtubules. This occurred at the level of increased localization, rather than increased total amounts, of the other kinetochore proteins. In contrast, in S. pombe, CENP-A overexpression resulted in increased numbers of CENP-A molecules per centromere, but not in increased recruitment of other kinetochore proteins, as measured by fluorescence microscopy [7]. C. albicans normally has 1 kinetochore-microtubule attachment per centromere core region of approximately 3kb, while S. pombe has 2–4 kinetochore-microtubule attachments within the centromere core region of approximately 4kb [7]. Therefore, in wild-type S. pombe, the density of kinetochore proteins is higher than in C. albicans and steric hindrance may prevent the formation of additional kinetochores.
Since each wild-type C. albicans centromere has only one kinetochore-microtubule attachment [7], the increase in kinetochore proteins and spindle microtubules could have several possible effects on chromosome segregation fidelity. Overexpression of CSE4 could increase chromosome segregation fidelity due to increases in the number of attachments between the centromere DNA and the spindle microtubules. Alternatively, the presence of extra kinetochore proteins could decrease chromosome segregation fidelity due to improper bundling of microtubules and the potential for merotelic attachments. To address the question of how increasing the number of spindle microtubules affects chromosome segregation fidelity, we measured the rate of loss of a heterozygous URA3 marker in non-inducing and inducing conditions for CSE4 overexpression. Interestingly, overexpression of CSE4 did not alter chromosome segregation fidelity (Figure 3D), suggesting that C. albicans kinetochore function is not limited by having increased numbers of microtubules. Previous work in S. cerevisiae found that the number of interpolar spindle microtubules can increase when cells are arrested in anaphase [26]. To ask if cells with overexpressed CSE4 have any cell cycle defects such as anaphase arrest, we calculated the percentage of cells with mitotic spindles in cells with normal and increased levels of Cse4 (Figure 3E). Importantly, no significant increase in mitotic cells was observed indicating that cells in which CSE4 is overexpressed do not have cell cycle delays. When centromeres are composed of >1 kinetochore-microtubule per centromere, they must be aligned to avoid inappropriate, merotelic associations. This function is accomplished by monopolin, a complex of proteins that ensures sister chromatid co-orientation during mitosis in S. pombe and during meiosis I in S. cerevisiae [19]. Because C. albicans chromosomes with increased spindle microtubules have no reduction in segregation fidelity, it appears that this function has been retained. Consistent with this, the C. albicans genome sequence includes ORFs predicted to encode monopolin complex members (orf19.7663 and orf19.3248) and these genes are expressed [27]. The result that extra microtubule attachments to do not alter chromosome segregation suggests that they retain function in aligning kinetochores in C. albicans and it is tempting to speculate that these ORFs have a role in that process.
The requirement for Dam1 proteins is reduced in strains with extra kinetochore components
Since the stoichiometry of kinetochore proteins and spindle microtubules at centromeres was increased in cells overexpressing CSE4 in C. albicans, we exploited this system to determine if the requirement for the Dam1 complex is reduced in cells with increased numbers of kinetochore and microtubule proteins. We combined the conditional overexpression of CSE4, under the control of the PCK1 promoter, with the regulated expression of DAD2 or DAM1, using the MET3 promoter. As a control, we used strains that overexpressed CSE4 and that retained an intact copy of the Dam1 complex gene in addition to one copy that was controlled by the MET3 promoter. Importantly, cells in which DAD2 or DAM1 expression was repressed and CSE4 was overexpressed grew significantly better (p<0.01) relative to the control strains than cells in which DAD2 or DAM1 expression was repressed and CSE4 was expressed at wild-type levels (Figure 4A). Furthermore, in DAM1 depletion conditions, the percentage of cells with normal chromosome segregation patterns increased from ~6% to approximately 36% when CSE4 was overexpressed (p<0.01) (Figure 4B). Given that ~60% of centromeres are estimated to have multiple kinetochore-microtubule attachments when CSE4 is overexpressed, this partial rescue of chromosome segregation is consistent with an increase in normal chromosome segregation at most, but not all, centromeres under these conditions.
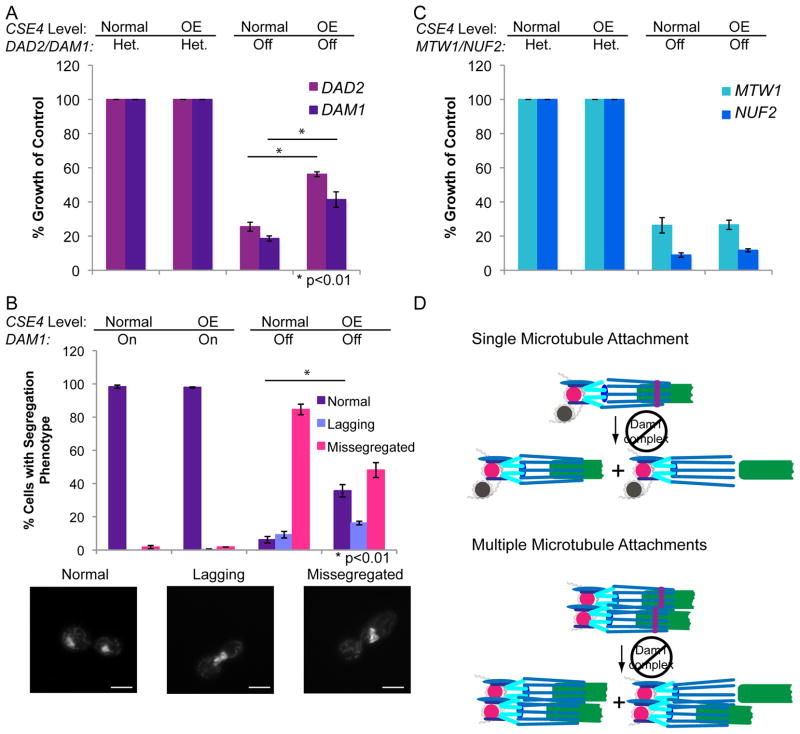
Figure 4. The requirement for Dam1 proteins is reduced in strains with extra kinetochore components.
(A) Relative colony size was used to quantify growth of CENP-A/PCK1p-CSE4 strains in which the only copy of DAD2 or DAM1 was controlled by the MET3 promoter (right columns) compared to strains with an intact DAD2 or DAM1 allele (Het.). Strains were grown on YPA-Glucose to repress the PCK1 promoter (normal expression) and MET3 promoters and on YPA-Succinate to activate the PCK1 promoter (overexpression – OE) and repress the MET3 promoter. Colony sizes were measured after 48 h incubation. Data shown are the mean ± SEM of 4 experiments. Differences between normal CSE4 expression and CSE4 overexpression were statistically significant (p<0.01) using a two-tailed unpaired Student’s t-test (*). (B) The CENP-A/PCK1p-CSE4 MET3p-DAM1/Δdam1 strain was grown in SDC-Glucose-Met-Cys to repress the PCK1 promoter and activate the MET3 promoter, SDC-Succinate-Met-Cys to activate the PCK1 and MET3 promoters, SDC-Glucose+Met+Cys to repress the PCK1 and MET3 promoters, or SDC-Succinate+Met+Cys to activate the PCK1 promoter and repress the MET3 promoter for 6 h. Repression of PCK1 results in normal levels of CSE4, while activation results in overexpression (OE). Cells were stained with DAPI, imaged at 1000x total magnification, and chromosome segregation was analyzed. Data shown are mean ± SEM of 3 experiments. Differences between normal CSE4 expression and CSE4 overexpression when DAM1 was repressed were statistically significant (p<0.01) (*) using a two-tailed unpaired Student’s t-test. Examples of each chromosome segregation pattern are below the figure. Scale bar = 2 μm. (C) Relative colony size was used to quantify growth of CENP-A/PCK1p-CSE4 strains in which the only copy of MTW1 or NUF2 was controlled by the MET3 promoter (right columns) compared to strains with an intact MTW1 or NUF2 allele in addition to the conditionally expressed allele. Strains were grown and colony size measured as in (A). Data shown are the mean ± SEM of 3 experiments. (D) Proposed mechanism to explain the reduced requirement for Dam1 proteins when the number of kinetochore-microtubule interactions is increased. Larger numbers of spindle microtubules increase the chances that a defective attachment can be reestablished, and thus reduce the requirement for the Dam1 processivity factor.
It was formally possible that depletion of the Dam1 complex caused major changes in kinetochore organization. To ask if this occurred, we examined the localization patterns of other kinetochore components, Mtw1-GFP (a MIND complex inner kinetochore component) and Nuf2-GFP (a member of the outer kinetochore Ndc80 complex), following DAM1 repression. Binding of Mtw1-GFP and Nuf2-GFP at the kinetochores was maintained when MET3p-DAM1 expression was repressed (Figure S4A). Thus, the assembly of the remainder of the kinetochore was not perturbed by repression of DAM1. Interestingly, kinetochore declustering, indicative of a loss of the binding between kinetochores and spindle microtubules [28], was observed for Mtw1-GFP and Nuf2-GFP localization following depletion of DAM1 (Figure S4A and Figure S4B). Importantly, this declustering of kinetochores was significantly reduced (p<0.01) following overexpression of CSE4 in the DAM1 depletion strains (Figure S4B). As a control to ensure that overexpression of CSE4 does not reduce the requirement for components of other essential kinetochore complexes, we compared the growth of strains conditionally expressing MTW1 or NUF2 to heterozygous strains that retained an intact copy of MTW1 or NUF2. In both cases, overexpression of CSE4 did not alter the requirement for MTW1 or NUF2 for growth (Figure 4C). Therefore, overexpression of CSE4 in C. albicans results in increased numbers of kinetochore proteins and spindle microtubule that partially bypass the requirement for components of the Dam1 complex (DAM1 and DAD2), but do not affect the requirement for essential components of the MIND and NDC80 complexes of the kinetochore
Conclusions
In this work, overexpression of CSE4 in C. albicans results in the recruitment of larger amounts of several kinetochore proteins and spindle microtubules and that these kinetochores with increased copy numbers are functional. While we have not conclusively shown that the extra kinetochore proteins are attached to the increased numbers of spindle microtubules, it is tempting to speculate that these are indeed extra kinetochore-microtubule attachments. Examination of the requirement of the Dam1 complex for viability when CSE4 is overexpressed suggests that the requirement of the Dam1 complex for viability is a function of whether an organism has a single or multiple kinetochore-microtubule attachments, rather than whether the organism has point or regional centromeres. In S. cerevisiae, the Dam1 complex has been proposed to help capture the depolymerization force of the spindle microtubule to help chromosomes segregate [29, 30]. The Dam1 complex has been proposed to form rings that surround the microtubule to couple the kinetochore to the microtubule [30, 31], although non-ring forms of the Dam1 complex may be sufficient to link kinetochores to spindle microtubules [32]. Our observation that increased numbers of kinetochore proteins and spindle microtubules reduce the requirement for the Dam1 complex is consistent with the previously published S. cerevisiae and S. pombe phenotypes and is also consistent with the role of the Dam1 complex as an in vivo processivity factor for the attachment of microtubules to kinetochore proteins such as Ndc80. Interestingly, the Dam1 complex components are present at a lower concentration per kinetochore in S. pombe than in S. cerevisiae or C. albicans [7]. Furthermore, the Dam1 complex is not evident in mammals or other higher eukaryotes, which also have multiple kinetochore-microtubule attachments. In humans, the Ska1 complex has been proposed to increase the stability of kinetochore-microtubule attachment [33, 34]. While the Ska1 complex has been proposed to be a functional homolog of the Dam1 complex in mammalian cells, the mechanism for the Ska1 complex is not clear and its dependence upon numbers of kinetochore-microtubules attachments has not been explored [35].
We propose that the Dam1 complex is required for viability in S. cerevisiae and C. albicans because its function as a processivity factor for kinetochore-microtubule binding is more critical when chromosome segregation is dependent upon a single kinetochore-microtubule. We suggest that when multiple microtubules are present, Ndc80 binding to kinetochore-microtubules is redundant, such that if a defect occurs at one kinetochore-microtubule, other kinetochore-microtubule attachments in the same centromere DNA region can help prevent the kinetochore and spindle microtubule from diffusing away from each other (Figure 4D). In summary, this study provides in vivo evidence, in an organism in which the number of kinetochore proteins and spindle microtubules at a regional centromere can be manipulated, that the Dam1 complex acts as a processivity factor to connect kinetochores to microtubules.
Supplementary Material
Acknowledgments
We would like to acknowledge Maryam Gerami-Nejad for assistance with strain construction and members of the Berman laboratory for many helpful discussions. We would also like to thank Meleah Hickman, Hung-Ji Tsai, Ben Harrison, Duncan Clarke, Tricia Davis, and members of the Davis laboratory for providing very useful feedback on the manuscript. This work is supported by a Ruth L. Kirschstein NRSA Individual Fellowship F32 AI800742 to L.S.B., funding from the University of Minnesota Undergraduate Research Opportunity Program to S.E.A., and by NIH/NIAID AI075096 to J.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li JM, Li Y, Elledge SJ. Genetic analysis of the kinetochore DASH complex reveals an antagonistic relationship with the ras/protein kinase A pathway and a novel subunit required for Ask1 association. Mol Cell Biol. 2005;25:767–778. doi: 10.1128/MCB.25.2.767-778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. Embo J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 9.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldo JT, Greagor SA, Iqbal AJ, Gittens AS, Grant KK. The Dad1 subunit of the yeast kinetochore Dam1 complex is an intrinsically disordered protein. Biochem Biophys Res Commun. 2010;400:313–317. doi: 10.1016/j.bbrc.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 12.Reedy JL, Floyd AM, Heitman J. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol. 2009;19:891–899. doi: 10.1016/j.cub.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol. 2003;50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 14.Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol. 2000;182:5730–5736. doi: 10.1128/jb.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanyal K, Carbon J. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc Natl Acad Sci U S A. 2002;99:12969–12974. doi: 10.1073/pnas.162488299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins KA, Castillo AR, Tatsutani SY, Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell. 2005;16:5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leuker CE, Sonneborn A, Delbruck S, Ernst JF. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene. 1997;192:235–240. doi: 10.1016/s0378-1119(97)00069-3. [DOI] [PubMed] [Google Scholar]
- 20.Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 24.Roy B, Burrack LS, Lone MA, Berman J, Sanyal K. CaMtw1, a member of the evolutionarily conserved Mis12 kinetochore protein family, is required for efficient inner kinetochore assembly in the pathogenic yeast Candida albicans. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07558.x. j.1365–2958.2011.07558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furniss K, Vas AC, Lane A, Clarke DJ. Assaying topoisomerase II checkpoints in yeast. Methods Mol Biol. 2009;582:167–187. doi: 10.1007/978-1-60761-340-4_14. [DOI] [PubMed] [Google Scholar]
- 26.O’Toole ET, Mastronarde DN, Giddings TH, Jr, Winey M, Burke DJ, McIntosh JR. Three-dimensional analysis and ultrastructural design of mitotic spindles from the cdc20 mutant of Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1–11. doi: 10.1091/mbc.8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cote P, Hogues H, Whiteway M. Transcriptional analysis of the Candida albicans cell cycle. Mol Biol Cell. 2009;20:3363–3373. doi: 10.1091/mbc.E09-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–4139. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grishchuk EL, Efremov AK, Volkov VA, Spiridonov IS, Gudimchuk N, Westermann S, Drubin D, Barnes G, McIntosh JR, Ataullakhanov FI. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc Natl Acad Sci U S A. 2008;105:15423–15428. doi: 10.1073/pnas.0807859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 32.Gao Q, Courtheoux T, Gachet Y, Tournier S, He X. A non-ring-like form of the Dam1 complex modulates microtubule dynamics in fission yeast. Proc Natl Acad Sci U S A. 2010;107:13330–13335. doi: 10.1073/pnas.1004887107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. The EMBO Journal. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, 3rd, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.