Figure 4. Maximum parsimony tree analysis of zebrafish Tbx22 within the eight major paralog subfamilies of T-box proteins.
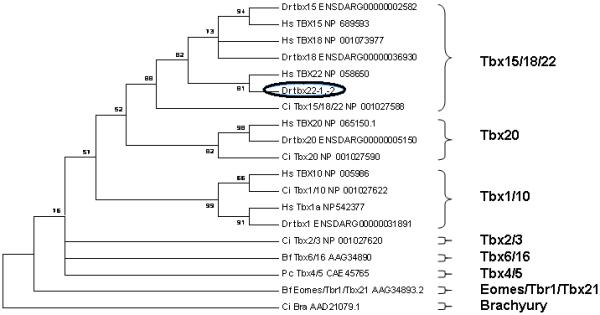
Paralogous zebrafish T-Box subfamily members found in the zebrafish ENSEMBL Zv7 assembly, release 46, were analyzed along with representative members of each paralog subfamily (see Supplemental Data File 1). Human TBX15, −18, −22, −1, −10, and −20 were included to provide more robust paralog family definitions. The maximum parsimony (MP) tree was constructed from the multi-sequence alignment of the 125 most conserved amino acids within the protein sequence (See Supplementary Data). This data is presented as a condensed, topology only maximum parsimony tree, rooted on the Ciona Brachyury T paralog group. Only bootstrap values above 50% are shown. The position of the zebrafish tbx22 is highlighted by a black oval.
