Fig. 5.
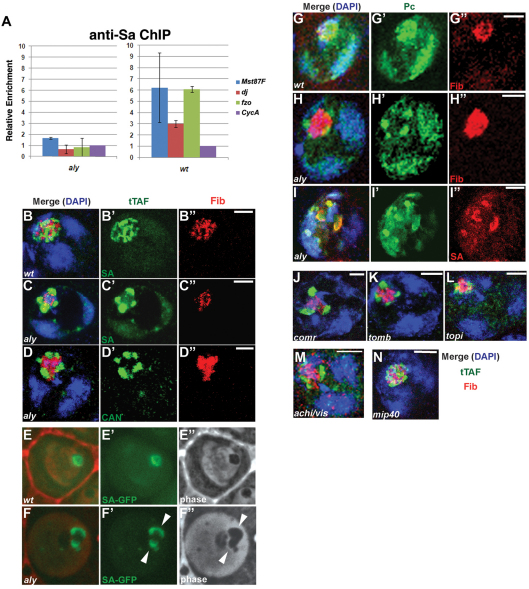
tMAC function is required for the recruitment of tTAFs to target genes and for proper subnuclear localization of tTAFs and Polycomb. (A) Real-time PCR analysis of ChIP using anti-Sa from aly and wild-type (wt) Drosophila testes. Relative enrichment by ChIP was computed as the ratio of ChIP DNA/input at the spermatid differentiation genes Mst87F, dj and fzo to ChIP DNA/input for the control gene CycA, which is expressed in both spermatogonia and spermatocytes independently of tMAC and tTAFs. Error bars indicate s.d. from three independent biological replicates). (B-I″) Localization of (B-F″) tTAFs and (G-I″) Polycomb (Pc) in wild-type and aly mutant spermatocytes by immunostaining of fixed cells (B-D″,G-I″) or in epifluorescent images of live cells (E-F″). Blue, DAPI; green, tTAF (Sa-GFP in B′,C′,E′,F′ or anti-Can in D′) or Pc-GFP (G′,H′,I′); red, the nucleolar marker anti-Fibrillarin (Fib in B″,C″,D″,G″,H″) or pseudo-color of the phase (E,F) or anti-Sa (I″). (J-N) Localization of Sa protein in spermatocytes mutant for other tMAC components: (J) comr, (K) tomb, (L) topi, (M) achi/vis, (N) mip40. Blue, DAPI; green, tTAF; red, anti-Fib. Scale bars: 4 μm.