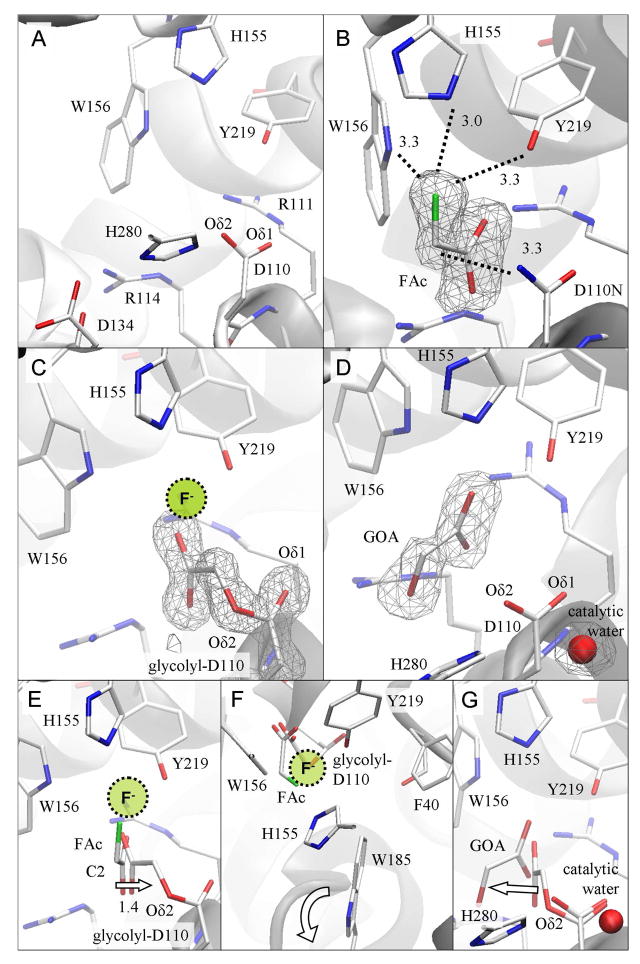
Figure 4. Structural comparison of reaction intermediates reveals the progress of enzymatic defluorination in RPA1163.
(A) Active site structure of the free WT FAcD. The cavity comprises the catalytic triad (Asp110-His280-Asp134), the carboxylate binding site (Arg111 and Arg114) and the fluoride pocket (His155, Trp156 and Tyr219). (B) The Asp110Asn/FAc Michaelis complex. The bound FAc reveals the key enzyme-substrate interactions (dashed lines with distances in Å). (C) The His280Asn/FAc covalent intermediate. The location where the displaced fluoride ion may be transiently bound is shown. (D) The WT/glycolate (GOA) product complex. (E) The initial SN2 attack is deduced from the superposition of FAc as found in the Michaelis complex onto the structure of the covalent intermediate. The arrow shows the displacement of the substrate’s C2 atom as this occurs. The location where the expelled fluoride ion may be transiently bound is indicated. (F) The same structural comparison viewed down the line of the atoms involved in the nucleophilic attack (i.e. from fluorine to the substrate’s C2 to Asp110’s Oδ2). The halide stabilizing residues (His155, Trp156 and Tyr219) form a ‘claw’ reaching for the leaving fluoride. Phe40 and Trp185 are also sufficiently close to participate in catalysis. The ‘flapping’ motion of the indole side chain of Trp185 is indicated by the curved arrow. (G) The subsequent hydrolysis of the covalent intermediate is inferred from the superposition of the glycolyl-Asp110 residue as found in the covalent intermediate onto the enzyme-product complex. This step employs the catalytic water activated by His280. The arrow indicates how the product glycolate (GOA) detaches from its former covalent bonding partner. The omit Fo−Fc electron density maps are contoured at 3 δ.