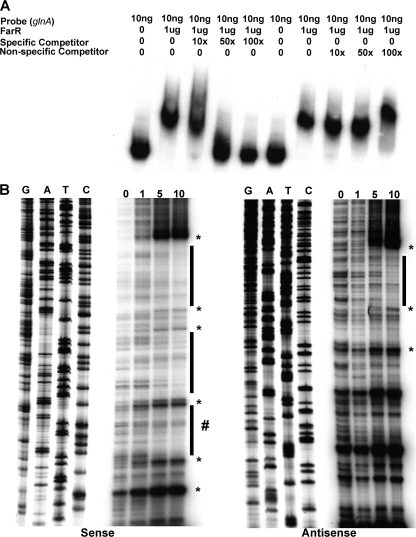
Fig. 4.
Identification of the FarR-binding site upstream of the glnA promoter. (A) The area of FarR-specific binding was first identified using the subsections of the full-length glnA sequence shown in Fig. 1. The full-length probe was PCR amplified using primers 5′pglnA and 3′pglnA, while binding site II was amplified with primers glnAsec2F and glnAsec2R. The nonspecific probe was prepared using primers rmpF and rmpR. FarR was found (data not presented) to bind only section II in a specific manner, as determined by competitive EMSA (shown in the figure). (B) The FarR-binding sites on the sense and antisense strands were identified by DNase I protection assays that employed increasing amounts of purified FarR-His (0, 1, 5, and 10 μg). The protected regions on each probe are identified by the black bars, with the protected region on the sense strand within section II being denoted by a # sign next to the bar to distinguish it from the other two protected regions that lie within section I. DNase I-hypersensitive sites are shown by an asterisk. The sequencing reactions for each probe are adjacent to the DNase I protection reactions and oriented G, A, T, C.