Figure 1.
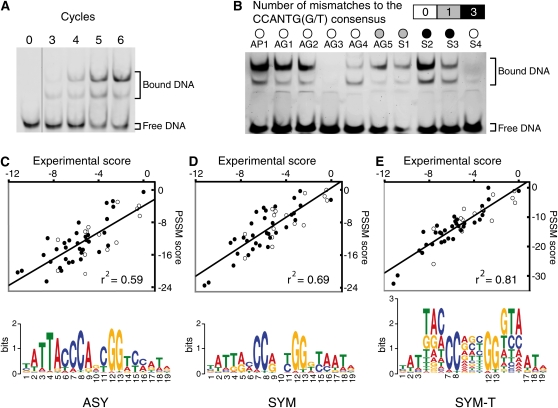
Optimization of the LFY Binding Site Model.
(A) Enrichment of DNA sequences bound by LFY over different Selex cycles.
(B) Binding of LFY to different sequences, either from AG or AP1 genes, or synthetic (S), with varying numbers of mismatches to the previously recognized consensus LFY binding motif.
(C) to (E) Comparison of experimentally determined and predicted scores (see Methods) for different DNA sequences with the three PSSMs (asymmetric [ASY], symmetric [SYM], and symmetric with triplets [SYM-T]), illustrated below by their logos. Open and closed circles represent sequences with or without the CCANTG[G/T] consensus, respectively.
[See online article for color version of this figure.]