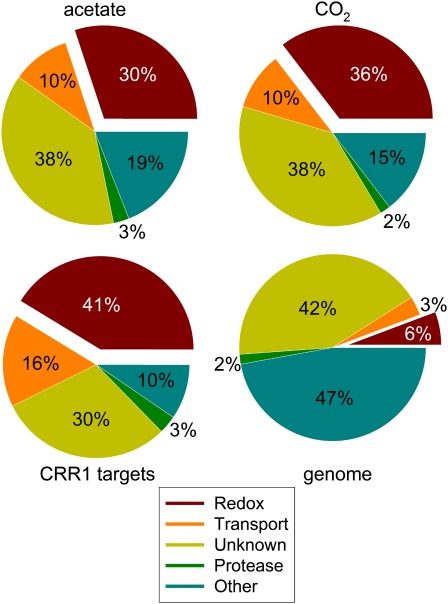
Figure 5.
Predicted Protein Domain Function for Copper Deficiency and CRR1 Targets.
The deduced protein sequences of the transcript set of 149 and 109 copper deficiency targets from Supplemental Data Set 3 online (acetate, photoheterotrophic; top left) and Supplemental Data Set 4 online (CO2, photoautotrophic; top right), respectively, were analyzed at the Pfam site (E-value ≤1 × 10−4). The identified protein domains were grouped according to their function into five categories: redox (red), transport (orange), protease (green), unknown (gold), and other (blue). Unknowns refer to protein sequences for which a domain could not be identified or if the domain was identified as having an unknown function (DUF). Domains that could not be classified as redox, transport, or protease were grouped into the “other” category. The distribution of protein domain functions in the subset of CRR1 targets (63 genes, bottom left) identified under photoheterotrophic conditions (see Figure 2A) is also shown. Chlamydomonas gene model set FM3.1 was used to estimate the genome-wide distribution of protein functions in the same categories. For the analysis of the complete genome (bottom right), the protein sequences with a predicted domain function with a bit score ≤10 were labeled unknown.