Figure 5. Complex ePMV scene rendered for different audiences and purposes.
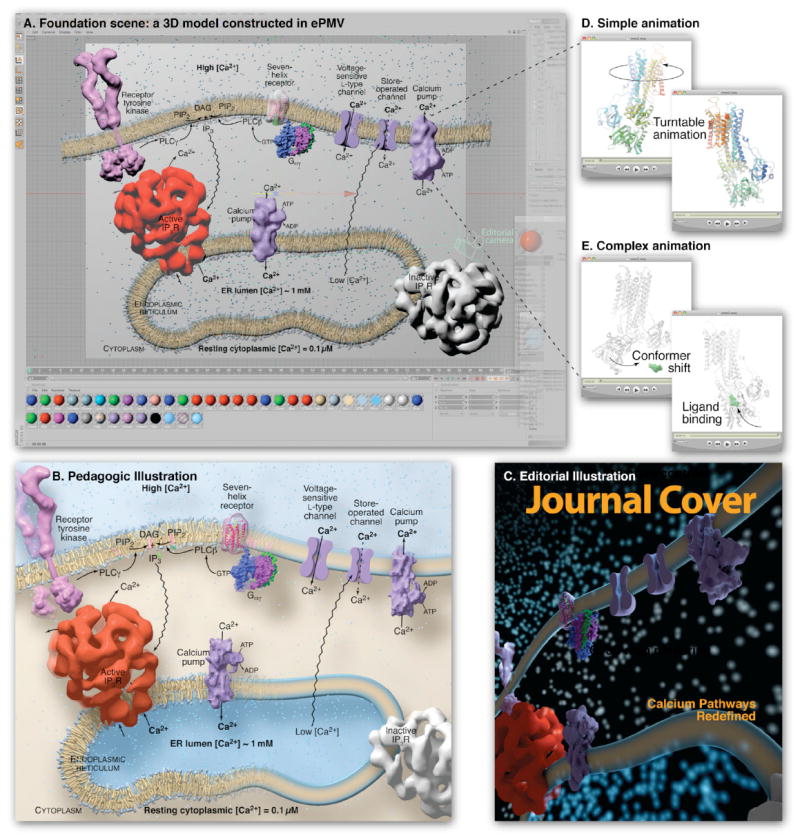
(A) A cell biology scene modeled with ePMV and host geometry, running in sample host Cinema 4D, depicts pathways of Ca2+ release and uptake using molecular structures from many sources. ePMV can quickly model representations of structural data while host tools can be used to model geometries to represent unknown or abstracted components. The host can then be used to assemble all of the models into a contextual scene. This scene includes a coarse molecular surface for receptor tyrosine kinase (PDB IDs 3kxz (Moy et al., 2010) and 2e9w (Yuzawa et al., 2007)) connected with two host cylinders to represent the unresolved transmembrane domains. Lipids are based on PDB coordinates pulled from a molecular dynamics simulation and simplified as an extruded rectangle on the right. The receptor and trimeric G protein, and calcium pump (entries 1f88; Palczewski et al. [2000]; and 1got; Lambright et al. [1996]) from the PDBTM, were modeled with coarse molecular surfaces (Bajaj et al., 1996) and the receptor made transparent using a host texture to reveal an underlying ePMV ribbon model. The calcium pump (PDB ID 1t5s (Sorensen et al.,2004)) was loaded from the optional Orientations of Protein Membranes Database to ensure a proper translation and procession in the membrane. Low-confidence representations of the voltage and store-operated channels were hand-modeled using host tools to complete the story with minimal distraction. The entry 1061 (Sato et al., 2004) was loaded from the EMDB (Henrick et al., 2003) and isocontoured (Bajaj et al., 1996) to represent the Active IP3 receptor and a gray texture was applied to a duplicated instance to represent the inactive state, since no density map models exist for this conformation. Please see http://epmv.scripps.edu/videos/structure2010 for a time-lapsed screen capture showing the construction of this model and a comparison to more traditional approaches. (B) A textbook figure based on the entire scene and rendered with educational style settings. (C) A second camera (“Editorial camera”) was placed at an extreme angle, and editorial style scene settings were selected to render the scene with a dramatic editorial mood. (D) A looping turntable animation of the calcium pump is useful for professional slide presentations and websites. (E) A detailed animation shows conformational change in the calcium pump, based on two crystallographic structures (PDB ID 1su4 (MacLennan and Green, 2000) and 1t5s (Sorensen et al.,2004)). Domain-localized linear interpolation of the atoms from a molecular dynamics simulation performed with Modeller in ePMV and a hinged bone skeleton (host) for large domain motion was used to morph between the two experimental conformations, host-based vibration was applied to the ligand to imply Brownian motion, and the position of the ligand was keyframed to migrate into the binding pocket at a specific time. A host physics engine was used to prevent the vibrating ligand from colliding with the atoms in the receptor.
