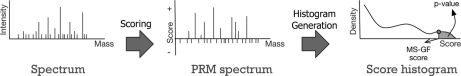
Fig. 1.
Computing p values with MS-GF for a single spectrum. Given a tandem mass spectrum, MS-GF converts the spectrum into a PRM spectrum (scored version of the tandem mass spectrum). The score of a PRM spectrum at mass m represents the log likelihood ratio that the peptide from which the spectrum was derived contains a prefix of mass m. Negative peaks in the PRM spectrum represent masses more likely to represent incorrect rather than correct prefix masses. Such negative peaks in the PRM spectrum usually correspond to low-intensity or missing peaks in the experimental spectrum. The PRM spectrum is used to compute the MS-GF score of any peptide against the spectrum. Then, MS-GF computes the histogram of the MS-GF scores of all peptides against the spectrum using the generating function approach. Finally, MS-GF computes the p value of a peptide as the area under the histogram with MS-GF scores equal or larger than the MS-GF score of the peptide.