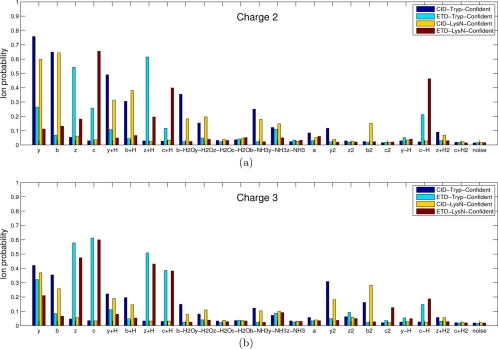
Fig. 4.
Probabilities of various ion types for the four types of (a) charge 2 spectra and (b) charge 3 spectra (see (32) for similar analysis). Spectra in CID-Tryp-Confident, ETD-Tryp-Confident, CID-LysN-Confident, and ETD-LysN-Confident were used. All the spectra were filtered to remove noisy peaks as follows: given a peak at mass M, we retained the peak if it is among the top six peaks within a window of size 100 Da around M. Precursor ions (or charge-reduced precursor ions) and their derivatives were also filtered out. A colored bar represents the probability (y axis) of a certain type of ion (x axis) being present in a filtered spectrum. Each data set is color coded. For example, a charge 2 spectrum in CID-Tryp-Confident generated from a length 10 peptide is expected to have 10–1 (number of potential cleavage sites) × 0.76 (probability of y ion) = 6.8 y ions, whereas a charge 2 spectrum in ETD-Tryp-Confident is expected to have only 9 × 0.26 = 2.3 y ions. In MS-GFDB, all ion types with probabilities exceeding 0.15 are used for scoring (see Supplement 1 for details).