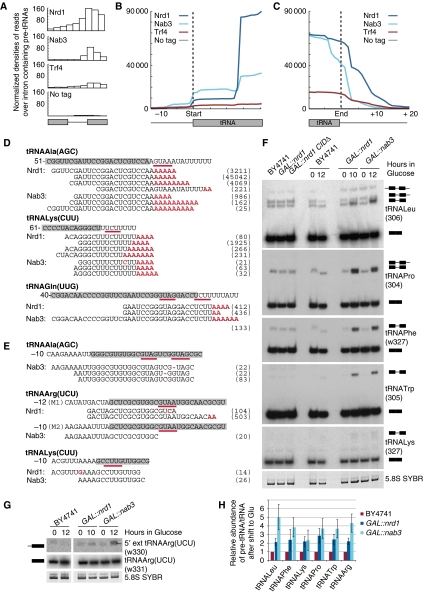
Figure 7.
Pre-tRNAs are targets for the nuclear RNA surveillance machinery. (A) Average densities of reads mapped to intron-containing tRNAs. tRNA exons and introns have various lengths; all exons and introns were divided into three bins and density of reads in each bin is displayed. (B, C) Average densities of reads of all tRNAs associated with the indicated proteins are plotted with respect to start (B) and end of the tRNA (C). (D, E) Alignments of high-throughput sequencing reads of RNAs associated with the indicated proteins to 3′ extended pre-tRNAs (D) and 5′ extended pre-tRNAs (E). Grey boxes mark mature tRNA sequences and numbering indicates nucleotide positions with respect to nucleotide +1 of the tRNA. Mismatches and deletions in sequencing reads are displayed in red. Nrd1 and Nab3 consensus-binding motifs are underlined. Numbers in brackets indicate the frequency with which each specific sequence was recovered in reads per million mapped sequences. (F, G) Northern analyses of total RNA from BY4741, GAL∷nrd1CIDΔ, GAL∷nrd1 and GAL∷nab3 strains. Oligonucleotide probes are given with the probe number in brackets. A schematic representation of the identified species is shown. (H) Quantification of pre-tRNA relative to mature tRNA is shown; expression at 12 versus 0 h is set to 1 for the WT. Average of three biological replicates is shown with s.d. See also Supplementary Figure S6.