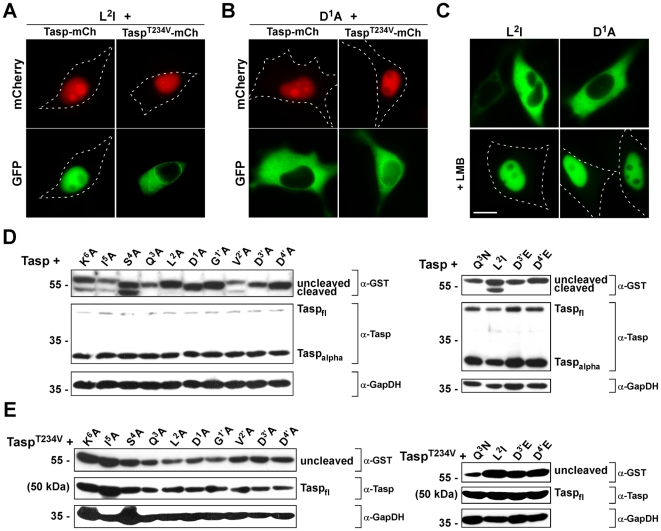
Figure 5. Identification of residues required for productive Taspase1 cleavage in living cells.
A.–C. Nuclear translocation of the indicated biosensor cleavage site mutants (TS-Cl2+CSmut) was analyzed in HeLa transfectants coexpressing the indicated biosensors together with Tasp- or inactive TaspT234V-mCherry. At least 200 fluorescent living cells were inspected, and representative examples are shown. Whereas substitution of Leu2 with Ile did not affect cleavage (A.), exchange of Asp1 with Ala completely abrogated cleavage (B.) LMB treatment verified that nuclear import of the variants was not affected. (C.) Scale bars, 10 µm. D./E. Cleavage of indicated cleavage site mutants by Tasp- (D.) or inactive TaspT234V-mCh (E.) analyzed by immunoblot. Notably, D1′A, D3′A and D4′A mutants run lower in the gel, most likely due to the loss of the negative charge. Expression of Taspase1 proteins as well as of cleavage products in 293T cell lysates was visualized using α-GST or -Taspase1 Abs. GAPDH served as loading control.