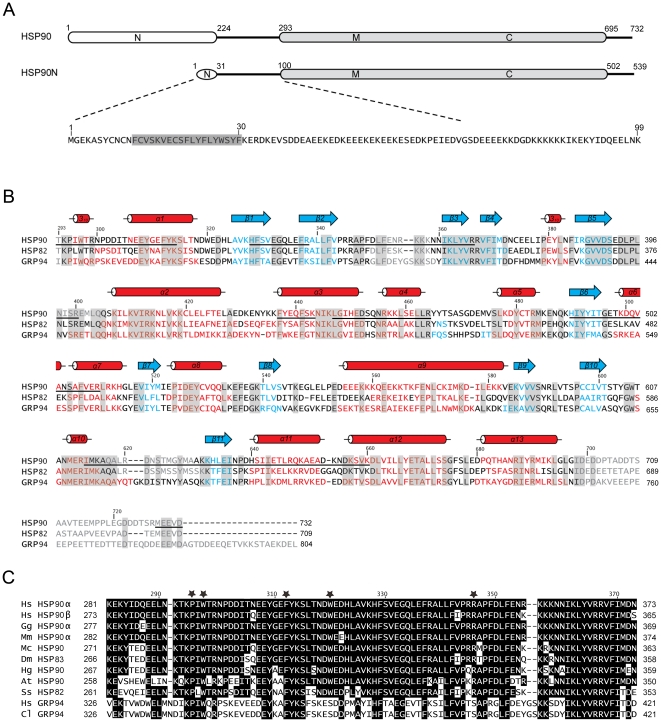
Figure 4. Sequence alignment.
(A) Schematic representation of human HSP90 and HSP90N. The functional domains (N, M, and C) and the residue number are indicated. The N-terminal sequence (residues 1–99) of HSP90N is shown below and a predicted transmembrane helix is highlighted in gray. (B) Structure-based sequence alignment of the MC domains of human HSP90, yeast HSP82 (PDB: 2CG9) and canine GRP94 (PDB: 2O1V). Residues of α-helices, β-strands, and disorders are shown in red, blue, and gray, respectively, and those of identical residues are highlighted by gray boxes. The secondary structural elements according to the human HSP90 structure are shown above the sequences, and the bars under the HSP90 sequence indicate the segments which have been identified by MALDI-MS analysis. Two extra Ala residues at the N-terminus and the C-terminal His-tag also have been identified, but are not shown in the figure. (C) Part of the M domain sequence from human HSP90 is aligned with other 90-kDa heat shock proteins of Homo sapiens (Hs), Gallus gallus (Gg), Mus musculus (Mm), Macrocentrus cingulum (Mc), Drosophila melanogaster (Dm), Heterodera glycines (Hg), Arabidopsis thaliana (At), Saccharomyce scerevisiae (Ss) and Canis lupus (Cl) for the regions near the W320 binding site. The residues involved in M domain interaction in the human MC-HSP90 hexamer are marked with black stars.