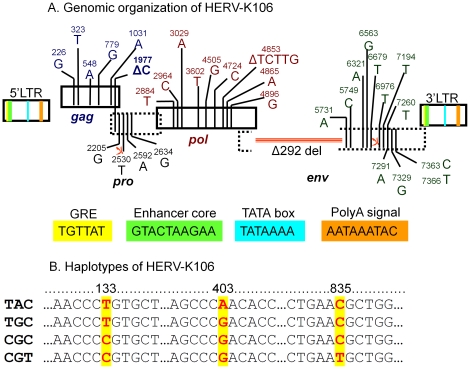
Figure 2. Genome organization and haplotypes of HERV-K106.
Figure 2A: Genomic characterization of HERV-K106 demonstrating the two LTRs, gag, pol, and env genes. The HERV-K106 genome was annotated with the aid of the HERV-K consensus sequence (HERV-KCON) [23], two HERV-K HML2 members that have somewhat functional gag (HERV-K101 and HERV-K109) [43], and insertionally polymorphic HERV-K113 that has intact ORF [17]. Known functional elements within the HERV-K106 LTR are shown in colored boxes. All SNP positions in the coding genes are counted from the beginning of gag ORF. Five nonsynonymous SNPs shown in gag region are HERV-K106 specific. These SNPs do not include I516M mutation which singlehandedly eliminates functionality of HERV-K113 gag [43] warranting the future investigation of functionality of K106 gag. Although K106 lacks this vital SNP it harbors one base deletion at position 1977 which causes a frameshift near the end of the gag gene. Whether this frameshift causes gag to be dysfunctional is unknown. The HERV-K106 polymerase gene (pol) that includes reverse transcriptase is distinct from the other HERV-K family members used in our sequence comparison. In addition to the SNP shown, it harbors a 5 bp deletion from 4849–4853 bp that results in a frameshift mutation. The large 292 base deletion beginning at 5392 bp of pol extending into the env gene is the signature of type-I HERV-K [16]. In addition to the 292 bp deletion, HERV-K106 env gene has a premature stop codon. (<$>\raster(70%)="rg1"<$>) indicate stop codons in HERV-K106 genome. Figure 2B: SNPs and surrounding bases in 3′LTR of HERV-K106 demonstrating four haplotypes. Each haplotype is listed on the left, each SNP is represented in red and the position containing the SNP is highlighted in yellow.