Abstract
Lipoprotein lipase (LIPL or LPL; E.C.3.1.1.34) serves a dual function as a triglyceride lipase of circulating chylomicrons and very-low-density lipoproteins (VLDL) and facilitates receptor-mediated lipoprotein uptake into heart, muscle and adipose tissue. Comparative LPL amino acid sequences and protein structures and LPL gene locations were examined using data from several vertebrate genome projects. Mammalian LPL genes usually contained 9 coding exons on the positive strand. Vertebrate LPL sequences shared 58–99% identity as compared with 33–49% sequence identities with other vascular triglyceride lipases, hepatic lipase (HL) and endothelial lipase (EL). Two human LPL N-glycosylation sites were conserved among seven predicted sites for the vertebrate LPL sequences examined. Sequence alignments, key amino acid residues and conserved predicted secondary and tertiary structures were also studied. A CpG island was identified within the 5'-untranslated region of the human LPL gene which may contribute to the higher than average (x4.5 times) level of expression reported. Phylogenetic analyses examined the relationships and potential evolutionary origins of vertebrate lipase genes, LPL, LIPG (encoding EL) and LIPC (encoding HL) which suggested that these have been derived from gene duplication events of an ancestral neutral lipase gene, prior to the appearance of fish during vertebrate evolution. Comparative divergence rates for these vertebrate sequences indicated that LPL is evolving more slowly (2–3 times) than for LIPC and LIPG genes and proteins.
Keywords: Vertebrates, amino acid sequence, lipoprotein lipase, evolution, gene duplication
Introduction
Lipoprotein lipase (LPL or LIPL; E.C.3.1.1.34) is one of three members of the triglyceride lipase family that contributes to vascular lipoprotein degradation and plays major roles in hydrolyzing circulating chylomicrons and very-low-density lipoproteins (VLDL) and in facilitating receptor-mediated lipoprotein uptake into heart, muscle and adipose tissue of the body (Wion et al., 1987; Dichek et al., 1991; Benlian et al., 1996). Hepatic lipase (HL; gene LIPC; E.C. 3.1.1.3) also serves a dual role in triglyceride hydrolysis and in ligand-binding for receptor-mediated lipoprotein uptake into the liver (Martin et al., 1988; Datta et al., 1988; Cai et al., 1989; Holmes et al., 2011a) whereas endothelial lipase (EL; gene LIPG; E.C.3.1.1.3) functions in high density lipoprotein (HDL) hydrolysis in the body (Jaye et al., 1999; Hirata et al., 1999; Holmes et al, 2011b). These enzymes are members of the vascular lipase gene family which have significant sequence similarities (Hirata et al., 1999; Ma et al., 2003; Brown & Rader, 2007).
The gene encoding LPL (LPL or LIPL) is expressed in various cells and tissues of the body, including heart, muscle, adipose tissue, brain, macrophages, lung, lactating mammary gland and endothelial cells where the enzyme hydrolyzes triglycerides from chylomicrons and very-low-density lipoproteins (VLDL) (Wion et al., 1987; Dichek et al., 1991; Benlian et al., 1996; Su et al., 2004). Studies of Lpl−/Lpl− knock out mice have shown that LPL-deficiency causes severe hypertriglyceridemia, reduced high-density lipoprotein (HDL) levels and death within 18 hours of birth (Weinstock et al., 1995). Human clinical studies have also examined loss of function LPL mutations leading to familial chylomicronemia or hyperlipoproteinemia type I, a rare recessive disorder appearing in children and characterized by dramatically reduced HDL-cholesterol ratios and very high blood triglyceride levels (Amies et al., 1991; Faustinella et al., 1991; Mead et al., 2002). In addition, human LPL polymorphisms influence significantly a number of major diseases, including atherosclerosis (Reymer et al., 1995; Shimo-Nakanishi et al., 2001; Tsutsumi, 2003; Stein & Stein, 2003), atherosclerotic cerebral infarction (Xu et al., 2008), ischemic stroke (Zhao et al., 2003), coronary artery disease (Zhang et al., 1998; Spence et al., 2003), pre-eclampsia (Hubel et al., 1999; Zhang et al., 2006), Alzheimer's disease (Papassotiropoulos et al., 2005; Blain et al., 2006), ulcerative colitis (Kosaka et al., 2006), hypertension (Chen et al., 2005), diabetes (Ukkola et al., 2005) and obesity (Huang et al., 2006; Radha et al., 2007).
Structures of several vertebrate LPL genes have been determined, including human (Wion et al., 1987; Chuat et al., 1992), mouse (Zechner et al., 1991), rat (Brault et al., 1992; The MGC Project Team, 2004) and chicken (Cooper et al., 1992). Several LPL cDNA and amino acid sequences have also been reported for other vertebrates including gorilla (Gorilla gorilla) and rhesus monkey (Macaca mulatta) (Martinez et al., 2001), baboon (Papio anubis) (Cole & Hixson, 1995), pig (Sus scrofa) (Harbitz et al., 1991), cow (Bos taurus) (Senda et al., 1987), sheep (Ovis aries) (Edwards et al., 1993), cat (Felis catus) (Ginzinger et al., 1996), goat (Capra hercus) (Badaoui et al., 2007) and guinea pig (Cavia porcellus) (Enerbaeck et al., 1987) and fish species, sea bass (Dicentrarchus labrax) (Jose Ilbanez et al., 2008) and bream (Sparus aurata; Pagrus major) (Saera-Vila et al., 2005; Oku et al., 2006). LPL genes usually contain 9 exons of DNA encoding LPL sequences which may undergo exon shuffling generating several isoproteins in each case (Thierry-Mieg and Thierry-Mieg, 2006). Three dimensional studies of pancreatic lipase (LIPP) (Winkler et al., 1990; Bourne et al., 1994) and molecular modeling of human LPL (van Tilbeurgh et al., 1994) have enabled identification of three major structural domains for the mammalian neutral lipase family, including an N-terminal domain with a catalytic triad of serine, aspartate and histidine residues; a `lid' domain which covers the active site and contributes to the specificity for triglyceride and phosphoglyceride substrates; and a C-terminal or `plat' domain, which contributes to lipid binding and specificity. LPL is synthesized by the endoplasmic reticulum (ER) of parenchymal cells and sequentially processed by the Golgi and ER with the addition of carbohydrate (Ailhaud, 1990; Stins et al., 1993; Hata et al., 1993). LPL is also subject to proprotein convertase cleavage at a site in the `hinge' region separating the N- and C-terminal enzyme domains (Jin et al., 2005) and behaves as a homodimer with a proposed head-to-tail conformation (Murthy et al., 1996; Wong et al., 1997; Kobayashi et al., 2002). Following secretion, LPL binds to heparan sulfate proteoglycans on the endothelial surface by electrostatic charge effects onto the luminal surface of capillary endothelial cells and macrophages (reviewed by Tsutsumi, 2003).
This paper reports the predicted gene structures and amino acid sequences for several vertebrate LPL genes and proteins, the predicted secondary and tertiary structures for vertebrate LIPL enzymes, several potential sites for regulating human LPL gene expression and the structural, phylogenetic and evolutionary relationships for these genes and enzymes with those for human, mouse and rat lipase gene families.
Methods
Vertebrate LPL gene and protein identification
BLAST (Basic Local Alignment Search Tool) studies were undertaken using web tools from the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al, 1997). Protein BLAST analyses used vertebrate LPL amino acid sequences previously described (Table 1). Non-redundant protein sequence databases for several mammalian genomes were examined using the blastp algorithm, including human (Homo sapiens) (International Human Genome Sequencing Consortium, 2001); chimpanzee (Pan troglodytes) (Chimpanzee Genome Analysis Consortium, 2005); orangutan (Pongo abelii) (http://genome.wustl.edu) ; cow (Bos taurus) (Bovine Genome Project, 2008); horse (Equus caballus) (Horse Genome Project, 2008); mouse (Mus musculus) (Mouse Sequencing Consortium, 2002); rat (Rattus norvegicus) (Rat Genome Sequencing Consortium, 2004); opossum (Monodelphis domestica) (Mikkelsen et al., 2007); platypus (Ornithorhynchus anatinus) (Warren et al., 2008); frog (Xenopus tropicalis) (http://genome.jgipsf.org/Xentr3/Xentr3.home.html); stickleback (http://www.broadinstitute.org/models/stickleback) (Gasterosteus aculeatus); and seasquirt (Ciona intestinalis) (http://genome.jgi-psf.org/ciona4/ciona4.info.html). This procedure produced multiple BLAST `hits' for each of the protein databases which were individually examined and retained in FASTA format, and a record kept of the sequences for predicted mRNAs and encoded LPL-like proteins . These records were derived from annotated genomic sequences using the gene prediction method: GNOMON and predicted sequences with high similarity scores for human LPL. Predicted LPL-like protein sequences were obtained in each case and subjected to analyses of predicted protein and gene structures.
Table 1.
Vertebrate lipoprotein lipase (LPL) genes and proteins
| Lipoprotein Lipase Gene LPL | Species | RefSeq ID 1Ensembl (predicted) | GenBank ID | UNIPROT ID | Amino acids | Chromosome location | Exons (strand) | Gene Size bps | pI | Subunit MW | Signal Peptide (Cleavage site) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Homo sapiens | NM_000237.2 | BC011353 | P06858 | 475 | 8:19,841,232–19,864,008 | 9 (+ve) | 22,777 | 8.4 | 53,163 | 1–27 [AA-AD] |
| Chimpanzee | Pan troglodytes | 1XP_001149804.1 | 2 | 2 | 475 | 8:16,183,708–16,206,551 | 9 (+ve) | 22,844 | 8.5 | 53,162 | 1–27 [AA-AD] |
| Orangutan | Pongo abelii | 3 | 2 | 2 | 475 | 8:19,482,810–19,505,529 | 9 (+ve) | 22,720 | 8.5 | 53,133 | 1–27 [AA-AD] |
| Rhesus | Macaca mulatta | 1ENSMMUT00000006658 | AF403770 | Q95MH0 | 475 | 8:19,847,146–19,860,974 | 9 (+ve) | 13,829 | 8.5 | 53,146 | 1–20 [TA-SR] |
| Baboon | Papio anubis | NP_001106082.1 | U18091 | P49060 | 475 | 2 | 2 | 2 | 8.5 | 53,146 | 1–20 [TA-SR] |
| Marmoset | Callithrix jacchus | 3 | 2 | 2 | 475 | Contig4830:148,445–170,321 | 9 (−ve) | 21,877 | 8.5 | 53,165 | 1–27 [DA-AD] |
| Mouse | Mus musculus | NM_008509.2 | BC003305 | P11152 | 474 | 8:71,404,652–71,426,282 | 9 (+ve) | 21,631 | 8.0 | 53,109 | 1–27 [AA-AD] |
| Rat | Rattus norvegicus | NP_036730.1 | BC081836 | Q06000 | 474 | 16:22,536,120–22,556,716 | 9 (−ve) | 20,597 | 8.4 | 53,082 | 1–27 [AA-AD] |
| Guinea Pig | Cavia porcellus | 1ENSCPOT00000004098 | 2 | P11153 | 475 | 1:57,048,993–57,068,443 | 9 (+ve) | 19,451 | 8.8 | 53,522 | 1–27 [AA-AK] |
| Horse | Equus caballus | 1XP_001489627.1 | 2 | 2 | 468 | 2:49,071,398–49,090,148 | 9 (+ve) | 18,751 | 9.0 | 52,467 | 1–21 [AA-DR] |
| Cow | Bos taurus | NP_001068588.1 | 2 | 2 | 478 | 8:70,187,336–70,209,826 | 9 (+ve) | 22,491 | 8.8 | 53,378 | 1–23 [RG-GL] |
| Dog | Canis familaris | 1XP_534584.2 | 2 | 2 | 471 | 25:40,075,103–40,095,543 | 9 (−ve) | 20,441 | 8.5 | 52,559 | 1–21 [AA-AR] |
| Rabbit | Oryctolagus cuniculus | NM_001177330.1 | FJ429312 | 2 | 474 | 15:4,554,425–4,578,617 | 9 (+ve) | 24,193 | 8.2 | 52,977 | 1–20 [TA-SR] |
| Pig | Sus scrofa | 1ENSSSCT00000010522 | AK344311 | P11151 | 478 | 14:3,826,571–3,852,602 | 9 (+ve) | 26,032 | 8.6 | 53,498 | 1–26 [LA-TA] |
| Elephant | Loxodonta africana | 1ENSLAFT00000005641 | 2 | 2 | 472 | 22:17,198,146–17,219,228 | 9 (+ve) | 21,083 | 9.0 | 52,937 | 1–20 [PA-SH] |
| Opossum | Monodelphis domestica | 1XP_001381955.1 | 2 | 2 | 478 | 1:580,795,573–580,818,319 | 9 (+ve) | 22,747 | 8.6 | 53,362 | 1–21 [TS-TG] |
| Platypus | Ornithorhynchus anatinus | 1ENSOANT00000009473 | 2 | 2 | 476 | 5:4,223,188–4,247,644 | 9 (−ve) | 24,457 | 8.4 | 53,558 | 1–26 [AA-SD] |
| Chicken | Gallus gallus | NM_205282 | AB016987 | P11612 | 475 | Z:53,400,437–53,408,327 | 9 (−ve) | 7,891 | 8.5 | 53,636 | 1–25 [AG-SD] |
| Frog | Xenopus tropicalis | 1ENSXETT00000056503 | 2 | 2 | 466 | 4sc79:338,410–419,025 | 9 (−ve) | 80,616 | 8.2 | 53,153 | 1–18 [AT-KL] |
| Stickleback | Gasterosteus aculeatus | 1ENSGACT00000015067 | 2 | 2 | 514 | VIII:14,407,768–14,412,555 | 10 (−ve) | 4,788 | 8.2 | 58,052 | 1–23 {FS-SD] |
RefSeq: the reference amino acid sequence
predicted Ensembl amino acid sequence
not available
predicted Ensembl amino acid sequence
Contig refers to a DNA scaffold for sequencing analyses
GenBank IDs are derived NCBI http://www.ncbi.nlm.nih.gov/genbank/; Ensembl ID was derived from Ensembl genome database http://www.ensembl.org; UNIPROT refers to UniprotKB/Swiss-Prot IDs for individual acid lipases (see http://kr.expasy.org); bps refers to base pairs of nucleotide sequences; pI refers to theoretical isoelectric points; the number of coding exons are listed.
BLAT analyses were subsequently undertaken for each of the predicted LPL amino acid sequences using the UC Santa Cruz genome browser [http://genome.ucsc.edu/cgi-bin/hgBlat] (Kent et al. 2003) with the default settings to obtain the predicted locations for each of the mammalian LPL genes, including predicted exon boundary locations and gene sizes. BLAT analyses were similarly undertaken for other human lipase genes using previously reported sequences for encoded lipases in each case (see Table 1). Structures for human and mouse isoforms (splicing variants) were obtained using the AceView website to examine predicted gene and protein structures (Thierry-Mieg and Thierry-Mieg, 2006) (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html?human).
Predicted Structures and Properties of Vertebrate Lipoprotein Lipases
Predicted secondary and tertiary structures for human and other vertebrate LPL proteins were obtained using the PSIPRED v2.5 web site tools provided by Brunel University (McGuffin et al. 2000) [http://bioinf.cs.ucl.ac.uk/psipred/psiform.html] and the SWISS MODEL web tools [http://swissmodel.expasy.org/], respectively (Guex & Peitsch 1997; Kopp & Schwede 2004). The reported tertiary structure for human pancreatic lipase related protein 1 (PLR1) (Walker et al., 2010) served as the reference for the predicted human, opossum and frog LPL tertiary structures, with modeling ranges of residues 36 to 450, 38 to 453 and 23 to 438 respectively. Theoretical isoelectric points and molecular weights for vertebrate LPL proteins were obtained using Expasy web tools (http://au.expasy.org/tools/pi_tool.html). SignalP 3.0 web tools were used to predict the presence and location of signal peptide cleavage sites (http://www.cbs.dtu.dk/services/SignalP/) for each of the predicted vertebrate LPL sequences (Emanuelsson et al 2007). The NetNGlyc 1.0 Server was used to predict potential N-glycosylation sites for vertebrate LPL proteins (http://www.cbs.dtu.dk/services/NetNGlyc/).
Comparative Human and Mouse Lipoprotein Lipase Gene (LPL) Expression
The genome browser (http://genome.ucsc.edu) (Kent et al. 2003) was used to examine GNF Expression Atlas 2 data using various expression chips for human and mouse LPL genes (http://biogps.gnf.org) (Su et al, 2004). Gene array expression `heat maps' were examined for comparative gene expression levels among human and mouse tissues showing high (red); intermediate (black); and low (green) expression levels.
Phylogeny Studies and Sequence Divergence
Alignments of vertebrate lipoprotein lipase (LPL), hepatic lipase (HL), endothelial lipase (EL), pancreatic lipase (LIPP), pancreatic lipase related proteins 1 and 2 (PLR1 and PLR2) and hormone sensitive lipase (HSL) sequences were assembled using BioEdit v.5.0.1 and the default settings (Hall, 1999). Alignment ambiguous regions, including the amino and carboxyl termini, were excluded prior to phylogenetic analysis yielding alignments of 429 residues for comparisons of vertebrate LPL sequences with other vertebrate lipase sequences and the seasquirt (Ciona intestinalis) lipase sequence (see Table 1 and Supplementary Table 1). Evolutionary distances were calculated using the Kimura option (Kimura, 1983) in TREECON (Van de Peer & de Wachter, 1994). Phylogenetic trees were constructed from evolutionary distances using the neighbor-joining method (Saitou & Nei, 1987) and rooted with the seasquirt lipase sequence. Tree topology was reexamined by the boot-strap method (100 bootstraps were applied) of resampling and only values that were highly significant (≥90) are shown (Felsenstein, 1985).
Results and Discussion
Alignments of Vertebrate Lipoprotein Lipase (LPL) Amino Acid Sequences with Pancreatic Lipase-Like Sequences
The deduced amino acid sequences for opossum (Monodelphis domestica), frog (Xenopus tropicalis) and stickleback (Gasterosteus aculeatus) LPL are shown in Figure 1 together with previously reported sequences for human (Wion et al., 1987; Dichek et al., 1991), mouse (Zechner et al., 1991), chicken LPL (Cooper et al., 1992; Raisonnier et al., 1995), horse pancreatic lipase (LIPP) (Bourne et al., 1994), human pancreatic lipase related protein 1 (LPR1) and human pancreatic lipase related protein 2 (LPR2) (Giller et al., 1992) (Table 1 and Supplementary Table). Alignments of human and other vertebrate LPL sequences examined showed between 58–99% identities, suggesting that these are products of the same family of genes, whereas comparisons of sequence identities of vertebrate LPL proteins with human and mouse HL and EL, horse LIPP, human PLR1 and PLR2 exhibited lower levels of sequence identities: HL (41% and 44% respectively); EL (44% and 45% respectively); LIPP (24%); PLR1 (26%); and PLR2 (24%) indicating that these are members of distinct lipase families (Table 2). The amino acid sequences for mammalian and chicken LPL contained 474–478 residues whereas frog (Xenopus tropicalis) and stickleback (G. aculeatus) LPL contained 466 and 514 amino acids, respectively, with the latter having extended N- and C-terminal sequences (Figure 1). Previous three dimensional studies of horse pancreatic lipase (LIPP) (Bourne et al., 1994) and modeling studies of human LPL (van Tilbeurgh et al., 1994) have enabled predictions of key residues for these vertebrate LPL proteins (sequence numbers refer to human LPL). These included the catalytic triad for the active site (Ser159; Asp183; His266); the hydrophobic N-terminus signal peptides (see also Table 1) which facilitate enzyme secretion into the circulation system (Jin et al., 2003); disulfide bond forming residues (Cys54/Cys68; Cys243/Cys266; Cys291/Cys302; Cys305/Cys310; Cys445/Cys466) (the latter disulfide bond is apparently absent in the stickleback LPL sequence); the predicted `lid' region (244–265) which covers the active site and participates in lipid substrate binding in analogous lipases (Winkler et al., 1990; Bourne et al., 1994); and a predicted `hinge' region for vertebrate LPL, containing a proteolytic cleavage site for proprotein convertase (320Arg-321Ala-322Lys-323Arg) (Jin et al., 2003; 2005) (see Figure 1). Specific tyrosine residues predicted for nitration following lipopolysaccharide (LPS) challenging were identified (tyrosines 121, 127 and 314) which down-regulate LIPL activity and reduce triglyceride clearance from the body (Casanovas et al, 2009). These residues were conserved for all of the vertebrate LPL sequences examined (Figure 1) possibly because of this role in reducing LPL activity following LPS administration, resulting in `lipemia' and increased binding of triglyceride-rich lipoproteins with LPS which undergo clearance by the liver (Gouni et al, 1993). With the exception of the N-terminus signal peptides, all of these sequences were strictly conserved or underwent conservative substitutions which may reflect the essential nature of these residues in contributing to LPL structure and function. The N-terminal region (residues 1–32 for human LPL) however underwent major changes in the number and sequence of amino acid residues but retained a predicted signal peptide property in each case (Figure 1; Table 1). The horse LIPP, human PLR1 and PLR2 sequences shared the catalytic triad residues, four of the five disulfide bonds predicted for the vertebrate LPL sequences and an N-signal peptide sequence property however other sequences were distinct with only 25% identical residues observed for horse LIPP and human LPL.
Figure 1. Amino Acid Sequence Alignments for Vertebrate Lipoprotein Lipase, Horse Pancreatic Lipase and Human Pancreatic Lipase Related Protein 1 and 2 Sequences.
See Table 1 for sources of lipoprotein lipase (LIPL or LPL) and pancreatic lipase-like sequences: Hu-human LPL; Mo-mouse LPL; Op-opossum LPL; St-stickleback LPL; PL-horse pancreatic lipase (LIPP); R1-human pancreatic lipase related 1 protein (PLR1); R2-human pancreatic lipase related protein 2 (PLR2); * shows identical residues for lipase subunits; : similar alternate residues;. dissimilar alternate residues; residues involved in N-signal peptide formation are shown in red; human N-glycosylated and potential N-glycosylated Asn sites are in green bold; active site (A) triad residues Ser (S); Asp (D); and His (H) are in pink bold; predicted disulfide bond Cys residues are shown in blue bold (•); α-helix for horse LIPP or predicted for vertebrate LIPL is in shaded yellow; β-sheet for horse LIPP or predicted for vertebrate LIPL is in shaded grey; bold underlined font shows residues corresponding to known or predicted exon start sites; exon numbers refer to human LPL gene exons; #### refers to residues which correspond to the horse LIPP `lid' region; xxxxx refers to the `hinge' region for horse LIPP ^ refers to hydrophobic amino acids in the `plat' domain which are located near to the active site triad in the LIPE dimer model reported by Griffon et al (2009); TSP refers to thrombospondin 1-like sequence; Δ refers to amino acid substitutions reported for human LIPL that result in activity loss and associated lipoproteinemia.
Table 2.
Percentage identities for vertebrate lipoprotein lipase (LPL), human and mouse hepatic lipase (HL), endothelial lipase (EL), horse pancreatic lipase (LIPP), human pancreatic lipase related protein 1 (PLR1) and human pancreatic lipase related protein (PLR2) amino acid sequences
| Lipase Gene | Human LPL | Rhesus LPL | Mouse LPL | Rat LPL | Horse LPL | Cow LPL | Opossum LPL | Chicken LPL | Frog LPL | Stickleback LPL | Human HL | Mouse HL | Human EL | Mouse EL | Horse LIPP | Human PLR1 | Human PLR2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human LPL | 100 | 99 | 92 | 91 | 94 | 92 | 81 | 72 | 72 | 58 | 41 | 44 | 44 | 45 | 24 | 26 | 24 |
| Rhesus LPL | 99 | 100 | 92 | 92 | 94 | 92 | 82 | 72 | 72 | 58 | 43 | 44 | 44 | 44 | 24 | 26 | 25 |
| Mouse LPL | 92 | 92 | 100 | 96 | 91 | 89 | 80 | 71 | 71 | 57 | 42 | 43 | 45 | 43 | 25 | 25 | 24 |
| Rat LPL | 91 | 92 | 96 | 100 | 91 | 89 | 81 | 71 | 71 | 58 | 42 | 43 | 45 | 45 | 24 | 25 | 26 |
| Horse LPL | 94 | 94 | 91 | 91 | 100 | 91 | 81 | 73 | 71 | 59 | 42 | 43 | 45 | 46 | 24 | 25 | 24 |
| Cow LPL | 92 | 92 | 89 | 89 | 91 | 100 | 80 | 72 | 69 | 58 | 41 | 41 | 44 | 45 | 24 | 25 | 23 |
| Opossum LPL | 81 | 82 | 80 | 81 | 81 | 80 | 100 | 70 | 70 | 55 | 39 | 41 | 43 | 44 | 23 | 25 | 25 |
| Chicken LPL | 72 | 72 | 71 | 71 | 73 | 72 | 70 | 100 | 75 | 60 | 42 | 40 | 49 | 46 | 25 | 26 | 24 |
| Frog LPL | 72 | 72 | 71 | 71 | 71 | 69 | 70 | 75 | 100 | 60 | 40 | 41 | 46 | 45 | 25 | 27 | 26 |
| Stickleback LPL | 58 | 58 | 57 | 58 | 59 | 58 | 55 | 60 | 60 | 100 | 36 | 33 | 39 | 39 | 25 | 26 | 26 |
| Human HL | 41 | 43 | 42 | 42 | 42 | 41 | 39 | 42 | 40 | 36 | 100 | 74 | 38 | 37 | 25 | 26 | 27 |
| Mouse HL | 44 | 44 | 43 | 43 | 43 | 41 | 41 | 40 | 41 | 33 | 74 | 100 | 42 | 40 | 29 | 26 | 26 |
| Human EL | 44 | 44 | 45 | 45 | 45 | 44 | 43 | 49 | 46 | 39 | 38 | 42 | 100 | 80 | 25 | 27 | 25 |
| Mouse EL | 45 | 44 | 43 | 45 | 46 | 45 | 44 | 46 | 45 | 39 | 37 | 40 | 80 | 100 | 26 | 26 | 26 |
| Horse LIPP | 24 | 24 | 25 | 24 | 24 | 24 | 23 | 25 | 25 | 25 | 25 | 29 | 25 | 26 | 100 | 57 | 64 |
| Human PLR1 | 26 | 26 | 25 | 25 | 25 | 25 | 25 | 26 | 27 | 26 | 26 | 26 | 27 | 26 | 57 | 100 | 52 |
| Human PLR2 | 24 | 25 | 24 | 26 | 24 | 23 | 25 | 24 | 26 | 26 | 27 | 26 | 25 | 26 | 64 | 52 | 100 |
Numbers show the percentage of amino acid sequence identities. Numbers in bold show higher sequence identities for lipases from the same gene family.
Heparin binding sites have been previously shown to play key roles in binding LPL and related neutral vascular lipases, HL and EL, to heparin sulfate proteoglycans on the luminal side of endothelial cells (Hill et al., 1998; Sendak & Bensadoun, 1998). Figure 2 summarizes the comparative amino acid sequences for vertebrate LPL for several sites previously investigated by Beg and coworkers (1998). A non-basic amino acid region similar to type 1 repeats of thrombospondin (TSP) (Prater et al., 1991) and four basic amino acid clusters are compared for 14 vertebrate LPL sequences, including 2 fish species, zebrafish (Danio rerio) and stickleback (G. aculeatus). Human LPL contains a TSP sequence (residues 414–421: Phe-Ser-Trp-Ser-Asp-Trp-Trp-Ser) similar to the repeats found in thromobospondin 1 (TSP1) that mediates cell-to-cell and cell-to-matrix interactions (see Wolf et al, 1990). The first and second Trp residues in this sequence were retained for all vertebrate LPL sequences examined which is consistent with in vitro studies of Beg and coworkers (1998) for synthetic LPL peptides. Comparisons of the four basic amino acid clusters showed conservation for these sequences (human LPL numbers used): cluster 1 (residues 287–312) retained Arg288, Lys292, Lys297, Arg304, Lys305 and Arg307 for all of the vertebrate LPL sequences examined with the exception of Lys292, which was substituted by Arg for the fish sequences, and Arg304, substituted by Lys for opossum LPL; cluster 2 (residues 315–332) retained Lys317, Arg319, Lys321, Arg322, Lys325, Lys330 and Arg332, with the exception of chicken LPL (first Lys substituted by Arg) and the fish LPL sequences examined for the second Lys, which was substituted by Arg (D. rerio) or by Thr (G. aculeatus); cluster 3 (residues 173–177) retained Lys173, Lys174 and Arg177 with the exception of the fish LPL sequences for which the first Lys was substituted by His; and cluster 4 (residues 329–441) which retained Lys330, Arg332, Lys334,Lys440 and Lys441, with the exception of Lys330 which was substituted by Arg for mouse and chicken LPL sequences and of Lys440, which was substituted by Gln for the fish LPL sequences. Synthetic peptide heparin binding properties for these clusters (Beg et al, 1998) is consistent with these results with clusters 1, 3 and 2, showing the strongest binding in vitro, whereas cluster 4 did not bind to heparin, under the conditions used in their study. Figure 2 also describes comparative sequences for homologous regions for horse LIPP and for human PLR1 and PLR2 sequences, which predominantly lacked these identified heparin binding basic amino acid residues.
Figure 2. Amino Acid Sequence Alignments for Predicted Heparin Binding Sites for Vertebrate Lipoprotein Lipases and Homologous Sequences for Pancreatic Lipase-Like Proteins.
Basic amino acid and thrombospondin (TSP)-like clusters reported by Beg et al (1998) for human LPL; K-lysine residue; R-arginine residue; W-tryptophan residue; numbers refer to C-terminal residue for each of the clusters examined for the 14 vertebrate LPL sequences examined; for consensus sequences, X refers to any amino acid; LIPP refers to pancreatic lipase; PLR1 and PLR2 refer to pancreatic lipase related proteins 1 and 2.
One N-glycosylation site has been previously reported for human LPL at 70Asn-71His-72Thr and (Kobayashi et al, 1994) and a second site predicted at 386Asn-387-Lys-388Thr (van Tilbeurgh et al, 1994). A comparative analysis of potential N-glycosylation sites for vertebrate LPL (Table 3) has shown that there are 7 sites overall although only two of these have been predominantly retained for the 20 vertebrate LPL sequences examined (designated as sites 2 and 4) (with the exception of stickleback LPL which has lost site 2 but gained site 1 at 34Asn-35Thr-36Thr) (Table 3). It is apparent from the study by Kobayashi and coworkers (1994) of a human LPL Asn70 variant that this N-glycosylation site is essential for catalysis and secretion. A key role for the second predicted N-glycosylation site is also likely given the conservation of this site for all vertebrate LPL sequences examined (Table 3). Other vascular neutral lipases contained four N-glycosylation sites which play key roles and contribute to enzyme stability, secretion and catalytic activity: HL (hepatic lipase) (Wolle et al. 1993; Ben-Zeev et al. 1994) and EL (endothelial lipase) (Miller et al. 2004; Skropeta et al. 2007). A single N-glycosylation site was observed for horse LIPP at 425Asn-426Leu-427Thr which is consistent with a previous report (Bourne et al., 1994). This latter predicted N-glycosylation site was also observed for human PLR1 and PLR2 although the latter sequence exhibited a second predicted site at 353Asn-354Phe-355Thr.
Table 3.
Predicted N-glycosylation sites for vertebrate lipoprotein lipases
| Vertebrate | Species | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 | Site 6 | Site 7 | No of Sites |
|---|---|---|---|---|---|---|---|---|---|
| Human | Homo sapiens | 70NHS | 386NKT | 2 | |||||
| Chimpanzee | Pan troglodytes | 70NHS | 386NKT | 2 | |||||
| Orangutan | Pongo abelii | 70NHS | 386NKT | 2 | |||||
| Rhesus | Macaca mulatta | 70NHS | 386NKT | 2 | |||||
| Baboon | Papio anubis | 70NHS | 386NKT | 2 | |||||
| Marmoset | Callithrix jacchus | 70NHS | 386NKT | 2 | |||||
| Mouse | Mus musculus | 70NHS | 386NKT | 2 | |||||
| Rat | Rattus norvegicus | 70NHS | 386NKT | 411NDS | 3 | ||||
| Guinea Pig | Cavia porcellus | 70NHS | 386NNT | 2 | |||||
| Horse | Equus caballus | 63NQS | 379NKT | 2 | |||||
| Cow | Bos taurus | 73NHS | 389NKT | 2 | |||||
| Dog | Canis familaris | 66NHT | 382NKT | 2 | |||||
| Rabbit | Oryctolagus cuniculus | 69NHS | 385NKT | 471NKS | 3 | ||||
| Pig | Sus scrofa | 73NHS | 389NKT | 2 | |||||
| Elephant | Loxodonta africana | 70NYS | 386NKT | 2 | |||||
| Opossum | Monodelphis domestica | 73NHS | 389NKT | 452NIS | 3 | ||||
| Platypus | Ornithorhynchus anatinus | 71NHT | 387NKT | 2 | |||||
| Chicken | Gallus gallus | 70NHT | 354NVT | 386NKT | 3 | ||||
| Frog | Xenopus tropicalis | 60NHT | 344NLT | 376NKT | 3 | ||||
| Stickleback | Gasterosteus aculeatus | 34NTT | 407NTT | 2 |
Numbers refer to amino acids in the acid sequences, including N-asparagine; K-lysine; I-isoleucine; H-histidine; S-serine; T-threonine; Q-glutamine; D-aspartate; Y-tyrosine; and V-valine. Note that there are 7 potential sites identified, including 2 sites for human LPL (sites 2 and 4) (see Kobayashi et al, 1994; van Tilbeurgh et al, 1994). N-glycosylation sites were identified using the NetNGlyc 1.0 web server (http://www.cbs.dtu.dk/services/NetNGlyc/).
Predicted Secondary and Tertiary Structures for Vertebrate LPL
Predicted secondary structures for vertebrate LPL sequences were compared with the previously reported secondary structure for horse LIPP (Bourne et al., 1994) and for human PLR1 (Walker et al., 2010) (Figure 1). α-Helix and β-sheet structures for the vertebrate LPL sequences were similar for several regions with the horse LIPP secondary structures. Consistent structures were predicted near key residues or functional domains including the β-sheet and α-helix structures near the active site residues (human LPL numbers used) Ser159, Asp 183 and His269; the `lid' domain (residues 243–265); and the `hinge' region (residues 317–330) which concludes with a β-sheet: Lys326-Met327-Tyr328-Leu329-Lys330. Figure 3 describes predicted tertiary structures for human, opossum (Monodelphis domestica) and frog (Xenopus tropicalis) LPL protein sequences and shows significant similarities for these polypeptides with horse pancreatic lipase (LIPP) (Bourne et al., 1994). The three LPL and LIPP domains were readily apparent, including the N-terminal `lipase' domain with the active site triad residues buried under the `lid' domain observed for horse LIPP. The `lid' has been previously shown to contribute to the preference for triglyceride and phospholipid substrates of other vascular lipases (HL and EL) (Dugi et al., 1995; Kobayashi et al., 1996) and may play a major role in determining the preference for triglyceride rich lipoprotein LPL substrates. A `hinge' region was also observed for vertebrate LPL proteins, separating the `lipase' and `plat' domains, with the latter having a `sandwich-like' β-pleated sheet structure. The `plat' domain for HL and EL has been previously shown to be essential for binding these enzymes to lipoprotein micelles and also contributes to preferences in lipoprotein binding (Wong et al, 1991; reviewed in Griffon et al, 2009). Biochemical studies have also shown that LPL behaves as a dimer (Olivecrona & Bengtsson-Olivecrona, 1983). In addition, a proprotein convertase proteolytic cleavage site was observed at the `hinge' region (Arg319-X320-Lys321-Arg322) (Figure 2), which may result in partially cleaved dimeric LPL forms with reduced activities and unknown biochemical roles, similar to those observed for endothelial lipase (Griffon et al, 2009). Comparisons of amino acid sequences studies for other vertebrate LPL proteins suggest that these properties and key sequences are substantially retained for all of the sequences examined.
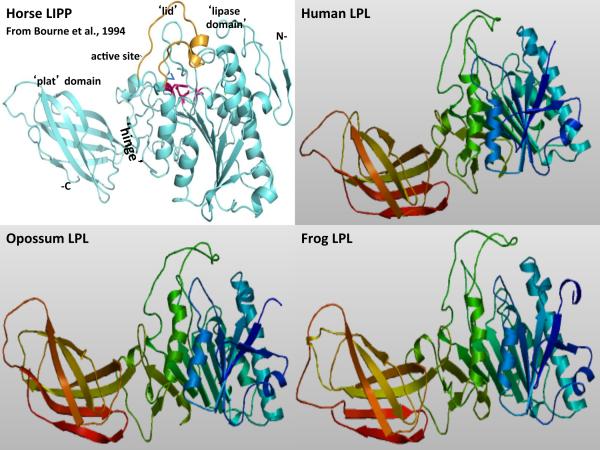
Figure 3. Tertiary Structure for Horse Lipoprotein Lipase and Predicted Tertiary Structures for Human, Opossum and Frog Lipoprotein Lipases.
The structure for horse pancreatic lipase (LIPP) is taken from Bourne et al, 1994; predicted human, opossum and frog lipoprotein lipase (LPL) tertiary structures were obtained using SWISS MODEL methods; the rainbow color code describes the tertiary structures from the N-(blue) to C-termini (red color) for human, opossum and zebrafish LPL; the horse LIPP tertiary structure shows the N- and C-termini, the `lipase', `lid' (in yellow) and `plat' domains which are separated by a `hinge' region; and the active site triad residues for horse LIPP which are shown in red.
Gene Locations and Exonic Structures for Vertebrate LPL Genes
Table 1 summarizes the predicted locations for vertebrate LPL genes based upon BLAT interrogations of several vertebrate genomes using the reported sequences for human (Wion et al., 1987; Chuat et al., 1992), mouse (Zechner et al., 1991), rat (Brault et al., 1992; The MGC Project Team, 2004) and chicken (Cooper et al., 1992) and the predicted sequences for other vertebrate LPL genes and the UC Santa Cruz genome browser (Kent et al. 2003). The predicted vertebrate LPL genes were predominantly transcribed on the positive strand, with the exception of the marmoset, rat, dog, platypus, chicken and frog genes, which were transcribed on the negative strand. Figure 1 summarizes the predicted exonic start sites for human, mouse, rat, opossum, chicken, frog and stickleback LPL genes with each having 9 coding exons, in identical or similar positions to those predicted for the human LPL gene (Wion et al., 1987; Chuat et al., 1992), with the exception of stickleback LPL, which contained an additional exon encoding an extended C-terminal sequence. Figure 4 shows the predicted structures of mRNAs for human and mouse LPL transcripts for the major isoform in each case (Theirry-Mieg & Thierry-Mieg, 2006). The transcripts were 27–28.5 kbs in length with 9 introns present for these LPL mRNA transcripts and in each case, an extended 3'-untranslated region (UTR) was observed. The human LPL genome sequence contained a CpG island (CpG112) which included the 5'-untranslated region of human LPL on chromosome 8. This CpG island within the LPL gene promoter may play a role in maintaining a very high level of gene expression (4.5 times the average for human genes) (Theirry-Mieg & Thierry-Mieg, 2006) which is similar to the CpG islands within housekeeping gene promoters expressed in most tissues (Saxonov et al., 2006).
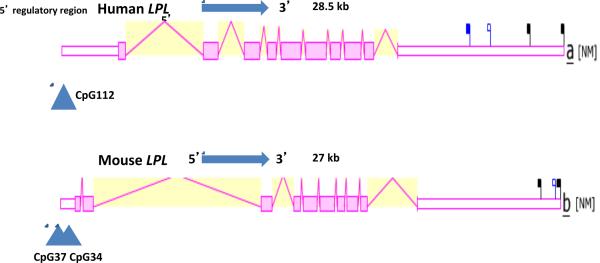
Figure 4. Gene Structures and Major Splicing Variant for the Human and Mouse Lipoprotein Lipase (LPL) Genes.
Derived from the AceView website http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/ (Thierry-Mieg and Thierry-Mieg, 2006); mature isoform variants (a) are shown with capped 5'-and 3'- ends for the predicted mRNA sequences; NM refers to the NCBI reference sequence; exons are in pink; the directions for transcription are shown as 5' → 3'; blue triangles show predicted CpG island sites at or near the 5'untranslated regions of the gene; the blue square shows a predicted microRNA binding site (miR29) observed at or near the human LPL 3'untranslated region; sizes of mRNA sequences are shown in kilobases (kb); predicted transcription factor binding sites (TFBS) for human LIPL are shown: FoxJ2-Foxhead J2 protein; Zic1-Zinc finger protein Zic1; E2F-transcription factor E2F2; HNF4 hepatocyte nuclear factor; ER-estrogen receptor DNA binding; PPARG-peroxisome proliferator-activated receptor γ.
Comparative Human and Mouse LPL Tissue Expression
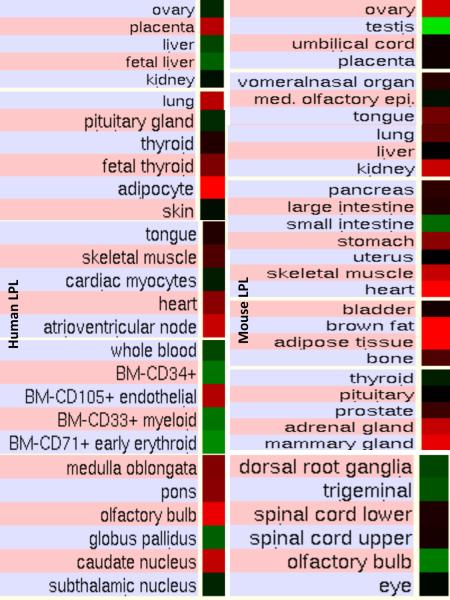
Figure 5 presents `heat maps' showing comparative gene expression for various human and mouse tissues obtained from GNF Expression Atlas Data using the U133A and GNF1H (human) and GNF1M (mouse) chips (http://genome.ucsc.edu; http://biogps.gnf.org) (Su et al, 2004). These data supported a broad and high level tissue expression for human and mouse LPL, particularly for heart, skeletal muscle, adipose tissue and lung, which is consistent with previous reports for these genes (Levak-Frank et al, 1999; Mead et al, 2002). Other comparisons of human and mouse LPL tissue expression indicated significant species differences, with higher levels of gene expression observed in human placenta and nerve tissues but lower expression levels in human liver, kidney and ovary than for the corresponding mouse tissues. Overall however, human and mouse LPL tissue expressions levels were >4 times the average level of gene expression which supports the key role played by this enzyme in lipid metabolism, especially in heart, skeletal muscle and adipose tissue (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/) (Theirry-Mieg & Thierry-Mieg, 2006).
Figure 5. Comparative Tissue Expression for Human and Mouse Lipoprotein Lipase Genes (LPL).
Expression `heat maps' (GNF Expression Atlas 2 data) (http://biogps.gnf.org) (Su et al, 2004) were examined for comparative gene expression levels among human and mouse tissues for LIPL genes showing high (red); intermediate (black); and low (green) expression levels. Derived from human and mouse genome browsers (http://genome.ucsc.edu) (Kent et al. 2003).
Phylogeny and Divergence of LPL and Other Vertebrate Lipase Sequences
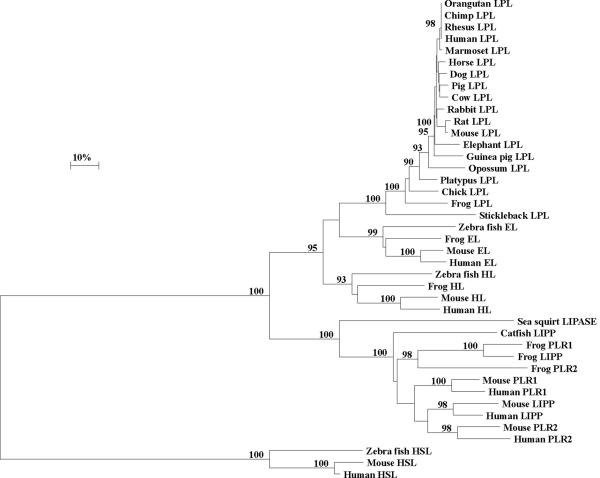
A phylogenetic tree (Figure 6) was calculated by the progressive alignment of 19 vertebrate LPL amino acid sequences with representative vertebrate hepatic lipase (HL), endothelial lipase (EL), pancreatic lipase (LIPP), pancreatic lipase related protein 1 (PLR1), pancreatic lipase related protein 2 (PLR2), hormone sensitive lipase (HSL) sequences and a Ciona intestinalis (sea squirt) lipase sequence (see Table 1 and Supplementary Table). The phylogram showed clustering of the LPL sequences into groups which were consistent with their evolutionary relatedness as well as groups for vertebrate HL and EL sequences (which formed a distinct cluster for the vascular lipases: LPL, EL and HL); for vertebrate LIPP, PLR1 and PLR2 (forming a pancreatic lipase-like cluster with the sea squirt lipase); and the vertebrate hormone sensitive lipases, which form a distinct cluster of sequences. These groups were significantly different from each other (with bootstrap values of ~ 100/100). It is apparent from this study of vertebrate LPL genes and proteins that this is an ancient protein for which a proposed common ancestor for the LIPC (encoding HL), LIPG (EL) and LPL (LPL) neutral lipase genes may have predated the appearance of fish during vertebrate evolution. This proposal is consistent with a previous report from Cohen (2003) which described predicted amino acid sequences for human and pufferfish (Takifugu rubripes) EL, LPL and HL. Genetic distances for human, cow, mouse and rat LPL, EL and HL sequences calculated from a mammalian common ancestor were as follows: 0.037±0.007, 0.086±0.007 and 0.125±0.011, respectively, which suggest that mammalian LPL sequences are diverging ~ 2–3 times more slowly than for HL and EL sequences. This is indicative of a conservative LPL protein during mammalian evolution.
Figure 6. Phylogenetic Tree of Vertebrate Lipoprotein Lipase (LPL) and other Vertebrate Lipase Amino Acid Sequences.
The tree is labeled with the lipase name and the name of the animal and is `rooted' with the seasquirt lipase sequence (Ciona intestinalis). Note the clusters corresponding to the LPL, LIPG and LIPC neutral lipase gene families, encoding lipoprotein lipase (LPL), endothelial lipase (EL) and hepatic lipase (HL), respectively; the pancreatic lipase-like gene cluster for the vertebrate LIPP, PLR1 and PLR2 genes encoding pancreatic lipase (LIPP), pancreatic lipase related protein 1 (PLR1) and pancreatic lipase related protein 2 (PLR2); and the hormone sensitive lipase (HSL) gene cluster for the vertebrate hormone sensitive lipases (HSL). A genetic distance scale is shown. The number of times a clade (sequences common to a node or branch) occurred in the bootstrap replicates are shown. Only replicate values of 90 or more which are highly significant are shown with 100 bootstrap replicates performed in each case.
Conclusions
The results of the present study indicate that vertebrate LPL genes and encoded enzymes represent a distinct gene and enzyme family of neutral lipases which share key conserved sequences that have been reported for other neutral lipases previously studied (Datta et al., 1988; Cai et al., 1989; Bourne et al., 1994; Jaye et al., 1999; Hirata et al., 1999; Holmes et al., 2011a, b). This enzyme has a unique property among the neutral lipases studied in hydrolyzing circulating chylomicrons and very low density lipoproteins (VLDL) and in facilitating receptor-mediated lipoprotein uptake into heart, muscle and adipose tissue of the body (Wion et al., 1987; Dichek et al., 1991; Benlian et al., 1996). LPL is encoded by a single gene among the vertebrate genomes studied which is highly expressed in human and mouse tissues, particularly in heart, adipose tissue and skeletal muscle, and usually contained 9 coding exons. Predicted secondary structures and tertiary structures for vertebrate LPL proteins showed a strong similarity with horse pancreatic lipase (LIPP) (Bourne et al., 1994). Three major structural domains were apparent for vertebrate LPL, including the `lipase' domain containing the catalytic triad residues; the `lid' which covers the active site and may contribute to the substrate specificities of neutral lipases (Dugi et al., 1995; Kobayashi et al., 1996); and the `plat' domain which contributes to lipoprotein binding (Wong et al., 1991). Phylogenetic studies using 19 vertebrate LPL with several representative hepatic lipase (HL), endothelial lipase (EL), pancreatic lipase-like (LIPP, PLR1 and PLR2) and hormone sensitive lipase (HSL) sequences indicated that the LPL gene has appeared early in vertebrate evolution, prior to the appearance of bony fish, and is evolving 2–3 times more slowly that the other vascular lipase genes and proteins (LIPG and LIPC) during vertebrate evolution.
Supplementary Material
RefSeq: the reference amino acid sequence; 1,3predicted Ensembl amino acid sequence; 2not available; 4Contig refers to a DNA scaffold for sequencing analyses; GenBank IDs are derived NCBI http://www.ncbi.nlm.nih.gov/genbank/; Ensembl ID was derived from Ensembl genome database http://www.ensembl.org; UNIPROT refers to UniprotKB/Swiss-Prot IDs for individual acid lipases (see http://kr.expasy.org).
Acknowledgements
This project was supported by NIH Grants P01 HL028972 and P51 RR013986. In addition, this investigation was conducted in facilities constructed with support from Research Facilities Improvement Program Grant Numbers 1 C06 RR13556, 1 C06 RR15456, 1 C06 RR017515. We also acknowledge the expert assistance of Dr Bharet Patel of Griffith University with the phylogeny studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ailhaud G. Cellular and secreted lipoprotein lipase revisited. Clin Biochem. 1990;23:343–347. doi: 10.1016/0009-9120(90)90034-r. [DOI] [PubMed] [Google Scholar]
- Altschul F, Vyas V, Cornfield A, Goodin S, Ravikumar TS, Rubin EH, Gupta E. Basic local alignment search tool. J Mol Biol. 1997;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ameis D, Kobayashi J, Davis RC, Ben-Zeev O, Malloy MJ, Kane JP, Lee G, Wong H, Havel RJ, Schotz MC. Familial chylomicronemia (type I hyperlipoproteinemia) due to a single missense mutation in the lipoprotein lipase gene. J. Clin Invest. 1991;87:1165–1170. doi: 10.1172/JCI115114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AshokKumar M, Subhashini NGV, Kanthimathi S, SaiBabu R, Ramesh A, Cherian KM, Emmanuel C. Associations for lipoprotein lipase and peroxisome proliferator-activated receptor-γ gene and coronary heart disease in an Indian population. Arch Med res. 2010;41:19–25. doi: 10.1016/j.arcmed.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Badaoui B, Serradilla JM, Tomas A, Urrutia B, Ares JL, Carrizosa J, Sanchez A, Jordana J, Amills M. Identification of two polymorphisms in the goat lipoprotein lipase gene and their association with milk production traits. J Dairy Sci. 2007;90:3012–3017. doi: 10.3168/jds.2006-409. [DOI] [PubMed] [Google Scholar]
- Beg OU, Uddin M, Siddiqi Analysis of heparin-binding sites in human lipoprotein lipase using synthetic peptides. J Prot Chem. 1998;17:807–815. doi: 10.1023/a:1020730418999. [DOI] [PubMed] [Google Scholar]
- Benlian P, De Gennes JL, Foubert L, Zhang H, Gagne SE, Hayden M. Premature atherosclerosis in patients with familial chylomicronemia caused by mutations in the lipoprotein lipase gene. N Engl J Med. 1996;335:848–854. doi: 10.1056/NEJM199609193351203. [DOI] [PubMed] [Google Scholar]
- Ben-Zeev O, Stahnke G, Liu RC, Davis RC, Doolittle MH. Lipoprotein lipase and hepatic lipase: the role of asparagines linked glycosylation in the expression of a functional enzyme. J Lipid Res. 1994;35:1511–1522. [PubMed] [Google Scholar]
- Blain JF, Aumont N, Theroux L, Dea D, Poirier J. A polymorphism in lipoprotein lipase affects the severity of Alzheimer's disease pathophysiology. Eur. J. Neurosci. 2006;24:1245–1251. doi: 10.1111/j.1460-9568.2006.05007.x. [DOI] [PubMed] [Google Scholar]
- Brault D, Noe L, Etienne J, Hamelin J, Raisonnier A, Souli A, Chuat J-C, Dugail I, Quignard-Boulange A, Lavau M, Galibert F. Sequence of rat lipoprotein lipase-encoding cDNA. Gene. 1992;121:237–246. doi: 10.1016/0378-1119(92)90127-b. [DOI] [PubMed] [Google Scholar]
- Brown RJ, Rader DJ. Lipases as modulators of atherosclerosis in murine models. Curr. Drug Targets. 2007;8:1307–1309. doi: 10.2174/138945007783220614. [DOI] [PubMed] [Google Scholar]
- Bourne Y, Martinez C, Kerfelec B, Lombardo D, Chapus C, Cambillau C. Horse pancreatic lipase. The crystal structure refined at 2.3-A resolution. J. Mol. Biol. 1994;238:709–732. doi: 10.1006/jmbi.1994.1331. [DOI] [PubMed] [Google Scholar]
- Bovine Genome Project 2008 http://hgsc.bcm.tmc.edu/projects/bovine.
- Cai SJ, Wong DM, Chen SH, Chan L. Structure of the human hepatic triglyceride lipase gene. Biochem. 1989;28:8966–8971. doi: 10.1021/bi00449a002. [DOI] [PubMed] [Google Scholar]
- Casanovas A, Carrascal M, Abián J, Lópes-Tejero MD, Llobera M. Lipoprotein lipase is nitrated in vivo after lipopolysaccharide challenge. Free Rad Biol Med. 2009;47:1553–1560. doi: 10.1016/j.freeradbiomed.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Chen P, Jou YS, Fann CS, Chen JW, Wu SY, Pan WH. Lipoprotein lipase gene is linked and associated with hypertension in Taiwan young-onset hypertension genetic study. J. Biomed. Sci. 2005;12:651–658. doi: 10.1007/s11373-005-7707-0. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Chuat J-C, Raisonnier A, Etienne J, Galibert F. The lipoprotein lipase-encoding human gene: sequence from intron-6 to intron-9 and presence in intron-7 of a 40-million-year-old Alu sequence. Gene. 1992;110:257–261. doi: 10.1016/0378-1119(92)90658-c. [DOI] [PubMed] [Google Scholar]
- Cohen JC. Endothelial lipase: direct evidence for a role in HDL metabolism. J Clin Invest. 2003;111:1–3. doi: 10.1172/JCI17744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SA, Hixson JE. Baboon lipoprotein lipase: cDNA sequence and variable tissue-specific expression of two transcripts. Gene. 1995;161:265–269. doi: 10.1016/0378-1119(95)00301-l. [DOI] [PubMed] [Google Scholar]
- Cooper DA, Lu SC, Viswanath R, Freiman RN, Bensadoun A. The structure and complete nucleotide sequence of the avian lipoprotein lipase gene. Biochim. Biophys. Acta. 1992;1129:166–171. doi: 10.1016/0167-4781(92)90482-f. [DOI] [PubMed] [Google Scholar]
- Datta S, Luo CC, Li WH, VanTuinen P, Ledbetter DH, Brown MA, Chen SH, Liu S, Chan L. Human hepatic lipase. Cloned cDNA sequence, restriction fragment length polymorphisms, chromosomal localization, and evolutionary relationships with lipoprotein lipase and pancreatic lipase. J Biol Chem. 1988;263:1107–1110. [PubMed] [Google Scholar]
- Dichek HL, Fojo SS, Beg OU, Skarlatos SI, Brunzell JD, Cutler GB, Jr, Brewer HB., Jr Identification of two separate allelic mutations in the lipoprotein lipase gene of a patient with the familial hyperchylomicronemia syndrome. J Biol Chem. 1991;266:473–477. [PubMed] [Google Scholar]
- Dugi KA, Dichek HL, Santamarina-Fojo S. Human hepatic and lipoprotein lipase: the loop covering the catalytic site mediates lipase substrate specificity. J. Biol. Chem. 1995;270:25396–25401. doi: 10.1074/jbc.270.43.25396. [DOI] [PubMed] [Google Scholar]
- Edwards WD, Daniels SW, Page RA, Volpe CP, Kille P, Sweeney GE, Cryer A. Cloning and sequencing of a full length cDNA encoding ovine lipoprotein lipase. Biochim Biophys Acta. 1993;1172:167–170. doi: 10.1016/0167-4781(93)90286-m. [DOI] [PubMed] [Google Scholar]
- Emmanuelsson O, Brunak S, von Heijne G, Nielson H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Enerbaeck S, Semb H, Bengtsson-Olivecrona G, Carlsson P, Hermansson M-L, Olivecrona T, Bjursell G. Molecular cloning and sequence analysis of cDNA encoding lipoprotein lipase of guinea pig. Gene. 1987;58:1–12. doi: 10.1016/0378-1119(87)90023-0. [DOI] [PubMed] [Google Scholar]
- Faustinella F, Chang A, van Biervliet JP, Rosseneu M, Vinaimont N, Smith LC, Chen S-H, Chan L. Catalytic triad residue mutation (Asp156-->Gly) causing familial lipoprotein lipase deficiency. Co-inheritance with a nonsense mutation (Ser447-->Ter) in a Turkish family. J Biol Chem. 1991;266:14418–14424. [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–789. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Giller T, Buchwald T, Blum-Kaelin D, Hunziger W. Two novel human pancreatic lipase related proteins, hPLRP1 and hPLRP2. Differences in colipase dependence and lipase activity. J Biol Chem. 1992;267:16509–16516. [PubMed] [Google Scholar]
- Ginzinger DG, Lewis MES, Ma Y, Jones BR, Liu G, Jones SD. A mutation in the lipoprotein lipase gene is the molecular basis of chylomicronemia in a colony of domestic cats. J Clin Invest. 1996;97:1257–1266. doi: 10.1172/JCI118541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffon N, Jin W, Petty DJ, Millar J, Badellino KO, Saven JG, Marchadier DH, Kempner ES, Billheimer J, Glick JM, Rader DJ. Identification of the active form of endothelial lipase, a homodimer in a head to tail conformation. J Biol Chem. 2009;284:23322–23330. doi: 10.1074/jbc.M109.037002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouni I, Oka K, Etienne J, Chan L. Endotoxin-induced hypertriglyceridemia is mediated by suppression of lipoprotein lipase at a post-transcriptional level. J Lipid Res. 1993;34:139–146. [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–99. [Google Scholar]
- Harbitz I, Kristensen T, Kran S, Davies W. Isolation and sequencing of porcine lipoprotein lipase cDNA and its use in multiallelic restriction fragment length polymorphism detection. Anim Genet. 1992;23:517–522. doi: 10.1111/j.1365-2052.1992.tb00170.x. [DOI] [PubMed] [Google Scholar]
- Hata A, Ridinger DN, Sutherland S, Emi M, Shuhua Z, Myers RL, Ren K, Cheng T, Inoue I, Wilson DE, Iverius P-H, Lalouel J-M. Binding of lipoprotein lipase to heparin. J Biol Chem. 1993;268:8447–8457. [PubMed] [Google Scholar]
- Hill JS, Davis RC, Yang D, Wen J, Philo JS, Poon PH, Phillips ML, Kempner ES, Wong H. Human hepatic lipase subunit structure determination. J Biol Chem. 1996;271:22931–22939. doi: 10.1074/jbc.271.37.22931. [DOI] [PubMed] [Google Scholar]
- Hirata K, Dichek HL, Cioffi JA, Choi SY, Leeper NJ, Quintana L, Kronmal GS, Cooper AD, Quertermous T. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J Biol Chem. 1999;274:14170–14175. doi: 10.1074/jbc.274.20.14170. [DOI] [PubMed] [Google Scholar]
- Holmes RS, VandeBerg JL, Cox LA. Vertebrate hepatic lipase genes and proteins: a review supported by bioinformatic studies. Open Access Bioinformatics. 2011a;3:1–11. doi: 10.2147/OAB.S18401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RS, VandeBerg JL, Cox LA. Vertebrate endothelial lipase: comparative studies of an ancient gene and protein in vertebrate evolution. Genetica. 2011b doi: 10.1007/s10709-011-9549-1. DOI 10.1007/s10709-011-9549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horse Genome Project 2008 http://www.uky.edu/Ag/Horsemap/
- Huang AQ, Hu YH, Zhan SY, Xu B, Pang ZC, Cao WH, Lu J, Qin Y, Lee LM. Lipoprotein lipase gene S447X polymorphism modulates the relation between central obesity and serum lipids, a twin study. Int J Obes. 2006;30:1693–1701. doi: 10.1038/sj.ijo.0803332. [DOI] [PubMed] [Google Scholar]
- Hubel CA, Roberts JM, Ferrell RE. Association of pre-eclampsia with common coding sequence variations in the lipoprotein lipase gene. Clin Genet. 1999;56:289–296. doi: 10.1034/j.1399-0004.1999.560406.x. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jaye M, Lynch KJ, Krawiec J, Marchadier D, Maugeais C, Doan K, South V, Amin D, Perrone M, Rader DJ. A novel endothelial-derived lipase that modulates HDL metabolism. Nat Genet. 1999;21:424–428. doi: 10.1038/7766. [DOI] [PubMed] [Google Scholar]
- Jin W, Sun G-S, Marchadier D, Octtaviani E, Glick JM, Rader DJ. Endothelial cells secrete triglyceride lipase and phospholipase activities in response to cytokines as a result of endothelial lipase. Circ Res. 2003;92:644–650. doi: 10.1161/01.RES.0000064502.47539.6D. [DOI] [PubMed] [Google Scholar]
- Jin W, Fuki IV, Seidah NG, Benjannet S, Glick JM, Rader DJ. Proprotein convertases are responsible for proteolysis and inactivation of endothelial lipase. J. Biol. Chem. 2005;280:36551–36559. doi: 10.1074/jbc.M502264200. [DOI] [PubMed] [Google Scholar]
- Jose Ibanez A, Peinado-Onsurbe J, Sanchez E, Cerda-Reverter JM, Prat F. Lipoprotein lipase (LPL) is highly expressed and active in the ovary of European sea bass (Dicentrarchus labrax L.) during gonadal development. Comp Biochem Physiol Part A Mol Integr Physiol. 2008;150:347–354. doi: 10.1016/j.cbpa.2008.04.598. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS. The human genome browser at UCSC. Genome Res. 2003;12:994–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge: 1983. [Google Scholar]
- Kobayashi J, Inadera H, Fujita Y, Talley G, Morisaki N, Yoshida S, Saito Y, Fojo SS, Brewer HB. A naturally occurring mutation at the second base of codon asparagine 43 in the proposed N-linked glycosylation site of human lipoprotein lipase: in vivo evidence that asparagine 43 is essential for catalysis and secretion. Biochem Biophys Res Commun. 1994;205:506–515. doi: 10.1006/bbrc.1994.2694. [DOI] [PubMed] [Google Scholar]
- Kobayashi J, Applebaum-Bowden D, Dugi KA, Brown DR, Kashyap VS, Parrott C, Duarte C, Maeda N, Santamarina-Fojo S. Analysis of protein structure-function in vivo. Adenovirus-mediated transfer of lipase lid mutants in hepatic lipase-deficient mice. J. Biol. Chem. 1996;271:26296–262301. [PubMed] [Google Scholar]
- Kobayashi Y, Nakajima T, Inoue I. Molecular modeling of the dimeric structure of human lipoprotein lipase and functional studies of the carboxyl-terminal domain. Eur J Biochem. 2002;269:4701–10. doi: 10.1046/j.1432-1033.2002.03179.x. [DOI] [PubMed] [Google Scholar]
- Kopp J, Schwede T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Yoshino J, Inui K, Wakabayashi T, Okushima K, Kobayashi T, Miyoshi H, Nakamura Y, Hayashi S, Shiraishi T, Watanabe M, Yamamoto T, Nakahara A, Katoh T. Impact of lipoprotein lipase gene polymorphisms on ulcerative colitis. World J Gastroenterol. 2006;12:6325–6330. doi: 10.3748/wjg.v12.i39.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levak-Frank S, Hofmann W, Weinstock PH, Radner H, Sattler W, Breslow JL, Zechner R. Induced mutant mouse lines that express lipoprotein lipase in cardiac muscle, but not in skeletal muscle and adipose tissue, have normal plasma triglyceride and high-density lipoproteincholesterol levels. Proc Natl Acad Sci USA. 1999;96:3165–3170. doi: 10.1073/pnas.96.6.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Cilingiroglu M, Otvos JD, Ballantyne CM, Marian AJ, Chan L. Endothelial lipase is a major genetic determinant for high-density lipoprotein concentration, structure and metabolism. Proc Natl Acad Sci USA. 2003;100:2748–2753. doi: 10.1073/pnas.0438039100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GA, Busch SJ, Meredith GD, Cardin AD, Blankenship DT, Mao SJT, Rechtin AE, Woods CW, Racke MM, Schafer MP, Fitzgerald MC, Burke DM, Flanagan MA, Jackson RL. Isolation and cDNA sequence of human postheparin plasma hepatic triglyceride lipase. J Biol Chem. 1988;263:10907–10914. [PubMed] [Google Scholar]
- Martinez J, Dugaiczyk LJ, Zielinski R, Dugaiczyk A. Human genetic disorders, a phylogenetic perspective. J Mol Biol. 2001;308:587–96. doi: 10.1006/jmbi.2001.4755. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: structure, function, regulation and role in disease. J. Mol. Med. 2002;80:753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res. 2002;43:1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, Jurka J, Kamal M, Mauceli E, Searle SMJ, Sharpe T, Baker ML, Batzer MA, Benos PV, Belov K, Clamp M, Cook A, Cuff J, Das R, Davidow L, Deakin JE, Fazzari MJ, Glass JL, Grabherr M, Greally JM, Gu W, Hore TA, Huttley GA, Kleber M, Jirtle RL, Koina E, Lee JT, Mahony S, Marra MA, Miller RD, Nicholls RD, Oda M, Papenfuss AT, Parra ZE, Pollock DD, Ray DA, Schein JE, Speed TP, Thompson K, VandeBerg JL, Wade CM, Walker JA, Waters PD, Webber C, Weidman JR, Xie X, Zody MC, Broad Institute Genome Sequencing Platform. Broad Institute Whole Genome Assembly Team. Marshall Graves JA, Ponting CP, Breen M, Samollow PB, Lander ES, Lindblad-Toh K. Genome of the marsupial Monodelphis domestica reveals innovation in noncoding sequences. Nature. 2007;447:167–175. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Miller GC, Long CJ, Bojilova ED, Marchadier D, Badellino KO, Blanchard N, Fuki IV, Glick JM, Rader DJ. Role of N-linked glycosylation in the secretion and activity of endothelial lipase. J Lipid Res. 2004;45:2080–2087. doi: 10.1194/jlr.M400162-JLR200. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Murthy V, Julien P, Gagné C. Molecular pathobiology of the human lipoprotein lipase gene. Pharmacol Therapeut. 1996;70:101–135. doi: 10.1016/0163-7258(96)00005-8. [DOI] [PubMed] [Google Scholar]
- Nagy L, Szanto A. Roles for lipid-activated transcription factors in atherosclerosis. Mol Nutr Food Res. 2005;49:1072–1074. doi: 10.1002/mnfr.200500097. [DOI] [PubMed] [Google Scholar]
- Oku H, Tokuda M, Okumura T, Umino T. Effects of insulin, triiodothyronine and fat soluble vitamins on adipocyte differentiation and LPL gene expression in the stromal-vascular cells of red sea bream, Pagrus major. Comp Biochem Physiol B Biochem Mol Biol. 2006;144:326–333. doi: 10.1016/j.cbpb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Olivecrona T, Bengtsson-Olivecrona G. Lipoprotein lipase from milk-the model enzyme in lipoprotein lipase research. In: Borensztajn J, editor. Lipoprotein Lipase. Evener; Chicago: 1987. pp. 15–58. [Google Scholar]
- Papassotiropoulos A, Wollmer MA, Tsolaki M, Brunner F, Molyva D, Lutjohann D, Nitsch RM, Hock C. A cluster of cholesterol-related genes confers susceptibility for Alzheimer's disease. J Clin Psychiatry. 2005;66:940–947. [PubMed] [Google Scholar]
- Prater CA, Plotkin J, Jaye D, Frazier WA. The properdin-like type I repeats of human thrombospondin contain a cell attachment site. J Cell Biol. 1991;112:1031–40. doi: 10.1083/jcb.112.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radha V, Vimaleswaran KS, Ayyappa KA, Mohan V. Association of lipoprotein lipase gene polymorphisms with obesity and type 2 diabetes in an Asian Indian population. Int J Obes. 2007;31:913–918. doi: 10.1038/sj.ijo.0803547. [DOI] [PubMed] [Google Scholar]
- Raisonnier A, Etienne J, Arnault F, Brault D, Noe L, Chuat J-C, Galibert F. Comparison of the cDNA and amino acid sequences of lipoprotein lipase in eight species. Comp Biochem Physiol B. 1995;111:385–398. doi: 10.1016/0305-0491(95)00006-t. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Reymer PWA, Gagne E, Groenemeyer BE, Zhang H, Forsyth I, Jansen H, Seidell JC, Kromhout D, Lie KE, Kastelein JJ, Hayden MR. A lipoprotein lipase mutation (Asn291Ser) is associated with reduced HDL cholesterol levels in premature atherosclerosis. Nat Genet. 1995;10:28–34. doi: 10.1038/ng0595-28. [DOI] [PubMed] [Google Scholar]
- Saere-Vila A, Calduch-Giner JA, Gomez-Requeni P, Medale F, Kaushik S, Perez-Sanchez J. Molecular characterization of gilthead sea bream (Sparus aurata) lipoprotein lipase. Transcriptional regulation by season and nutritional condition in skeletal muscle and fat storage tissues. Comp Biochem Physiol B. 2005;142:224–232. doi: 10.1016/j.cbpb.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–411. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda M, Oka K, Brown WV, Qasba PK, Furuichi Y. Molecular cloning and sequence of a cDNA coding for bovine lipoprotein lipase. Proc Natl Acad Sci USA. 1987;84:4369–4373. doi: 10.1073/pnas.84.13.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendak RA, Bensadoun A. Identification of a heparin-binding domain in the distal carboxyl-terminal region of lipoprotein lipase by site-directed mutagenesis. J Lipid Res. 1998;39:1310–1315. [PubMed] [Google Scholar]
- Shimo-Nakanishi Y, Urabe T, Hattori N, Watanabe Y, Nagao T, Yokochi M, Hamamoto M, Mizuno Y. Polymorphism of the lipoprotein lipase gene and risk of atherothrombotic cerebral infarction in the Japanese. Stroke. 2001;32:1481–1486. doi: 10.1161/01.str.32.7.1481. [DOI] [PubMed] [Google Scholar]
- Skropeta D, Settasatian C, McMahon MR, Shearston K, Caiazza D, McGrath KC, Jin W, Rader DJ, Barter PJ, Rye KA. N-Glycosylation regulates endothelial lipase-mediated phospholipid hydrolysis in apoE- and apoA-I-containing high density lipoproteins. J Lipid Res. 2007;48:2047–2057. doi: 10.1194/jlr.M700248-JLR200. [DOI] [PubMed] [Google Scholar]
- Spence JD, Ban MR, Hegele RA. Lipoprotein lipase (LPL) gene variation and progression of carotid artery plaque. Stroke. 2003;34:1176–1180. doi: 10.1161/01.STR.0000069160.05292.41. [DOI] [PubMed] [Google Scholar]
- Stein Y, Stein O. Lipoprotein lipase and atherosclerosis. Atherosclerosis. 2003;170:1–9. doi: 10.1016/s0021-9150(03)00014-5. [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Stins MF, Sivaram P, Sasaki A, Goldberg IJ. Specificity of lipoprotein lipase binding to endothelial cells. J Lipid Res. 1993;34:1853–1861. [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KR, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the human and mouse protein encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The MGC Project Team The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12. doi: 10.1186/gb-2006-7-s1-s12. http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html?human. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi K. Lipoprotein lipase and atherosclerosis. Current Vascular Pharmacol. 2003;1:11–17. doi: 10.2174/1570161033386673. [DOI] [PubMed] [Google Scholar]
- Ukkola O, Salonen J, Kesaniemi YA. Role of candidate genes in the lipid responses to intensified treatment in Type 2 diabetes. J. Endocrinol. Invest. 2005;28:871–875. doi: 10.1007/BF03345317. [DOI] [PubMed] [Google Scholar]
- Van De Peer Y, de Wachter R. TreeCon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Sci. 1994;10:569–575. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- van Tilbeurgh H, Roussel A, Lalouel J-M. Lipoprotein lipase. Molecular model based on the pancreatic lipase X-ray structure: consequences for heparin binding and catalysis. J Biol Chem. 1994;269:4626–4633. [PubMed] [Google Scholar]
- Walker JR, Davis T, Seitova A, Butler-Cole C, Weigelt J, Sundstrom M, Arrowsmith CH, Edwards AM, Bochkarev A, Dhe-Paganon S. Protein data bank entry for human pancreatic lipase-related protein. 2010;1(2PPL) http://www.rcsb.org/pdb/explore/explore.do?structureId=2PPL. [Google Scholar]
- Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grützner F, Belov K. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–83. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock PH, Bisgaier CL, Aalto-Setälä K, Radner H, Ramakrishnan R, Levak-Frank S, Essenburg AD, Zechner R, Breslow JL. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J Clin Invest. 1995;96:2555–2568. doi: 10.1172/JCI118319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler FK, D'Arcy A, Hunziker W. Structure of human pancreatic lipase. Nature. 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- Wion KL, Kirchgessner TG, Lusis AJ, Schotz MC, Lawn RM. Human lipoprotein lipase complementary DNA sequence. Science. 1987;235:1638–1641. doi: 10.1126/science.3823907. [DOI] [PubMed] [Google Scholar]
- Wolf FW, Eddy RL, Shows TB, Dixit VM. Structure and chromosomal localization of the human thrombospondin gene. Genomics. 1990;6:685–91. doi: 10.1016/0888-7543(90)90505-o. [DOI] [PubMed] [Google Scholar]
- Wolle J, Jansen H, Smith LC, Chan L. Functional role of N-linked glycosylation in human hepatic lipase: asparagine-56 is important for both enzyme activity and secretion. J. Lipid Res. 1993;34:2169–2175. [PubMed] [Google Scholar]
- Wong H, Davis RC, Nikazy J, Seebart KE, Schotz MC. Domain exchange: characterization of a chimeric lipase of hepatic lipase and lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 1991;88:11290–11294. doi: 10.1073/pnas.88.24.11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H, Hill JS, Davis RC, Nikazy J, Schotz MC. A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc Natl Acad Sci USA. 1997;94:5594–5598. doi: 10.1073/pnas.94.11.5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E, Li W, Zhan L, Guan G, Wang X, Chen S, Shi Y. Polymorphisms of the lipoprotein lipase gene are associated with atherosclerotic cerebral infarction in the Chinese. Neuroscience. 2008;155:403–408. doi: 10.1016/j.neuroscience.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Zechner R, Newman TC, Steiner E, Breslow JL. The structure of the mouse lipoprotein lipase gene: a B1 repetitive element is inserted into the 3' untranslated region of the mRNA. Genomics. 1991;11:62–76. doi: 10.1016/0888-7543(91)90102-k. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Liu Y, Liu BW, Fan P, Cavanna J, Galton DJ. Common genetic variants of lipoprotein lipase and apolipoproteins AI-CIII that relate to coronary artery disease: a study in Chinese and European subjects. Mol Genet Metab. 1998;64:177–183. doi: 10.1006/mgme.1998.2712. [DOI] [PubMed] [Google Scholar]
- Zhang C, Austin MA, Edwards KL, Farin FM, Li N, Hsu L, Srinouanprachanh SL, Williams MA. Functional variants of the lipoprotein lipase gene and the risk of preeclampsia among non-Hispanic Caucasian women. Clin Genet. 2006;69:33–39. doi: 10.1111/j.1399-0004.2005.00541.x. [DOI] [PubMed] [Google Scholar]
- Zhao S-P, Tong Q-G, Xiao Z-J, Cheng Y-C, Zhou H-N, Shai Nie S. The lipoprotein lipase Ser447Ter mutation and risk of stroke in the Chinese. Clin Chim Acta. 2003;330:161–164. doi: 10.1016/s0009-8981(03)00016-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RefSeq: the reference amino acid sequence; 1,3predicted Ensembl amino acid sequence; 2not available; 4Contig refers to a DNA scaffold for sequencing analyses; GenBank IDs are derived NCBI http://www.ncbi.nlm.nih.gov/genbank/; Ensembl ID was derived from Ensembl genome database http://www.ensembl.org; UNIPROT refers to UniprotKB/Swiss-Prot IDs for individual acid lipases (see http://kr.expasy.org).