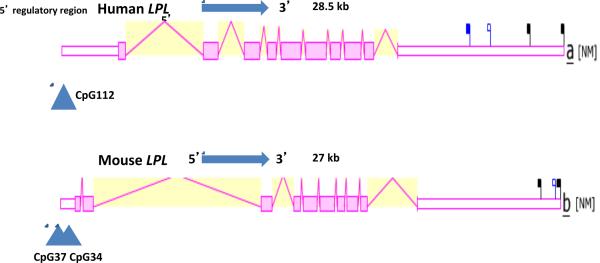
Figure 4. Gene Structures and Major Splicing Variant for the Human and Mouse Lipoprotein Lipase (LPL) Genes.
Derived from the AceView website http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/ (Thierry-Mieg and Thierry-Mieg, 2006); mature isoform variants (a) are shown with capped 5'-and 3'- ends for the predicted mRNA sequences; NM refers to the NCBI reference sequence; exons are in pink; the directions for transcription are shown as 5' → 3'; blue triangles show predicted CpG island sites at or near the 5'untranslated regions of the gene; the blue square shows a predicted microRNA binding site (miR29) observed at or near the human LPL 3'untranslated region; sizes of mRNA sequences are shown in kilobases (kb); predicted transcription factor binding sites (TFBS) for human LIPL are shown: FoxJ2-Foxhead J2 protein; Zic1-Zinc finger protein Zic1; E2F-transcription factor E2F2; HNF4 hepatocyte nuclear factor; ER-estrogen receptor DNA binding; PPARG-peroxisome proliferator-activated receptor γ.