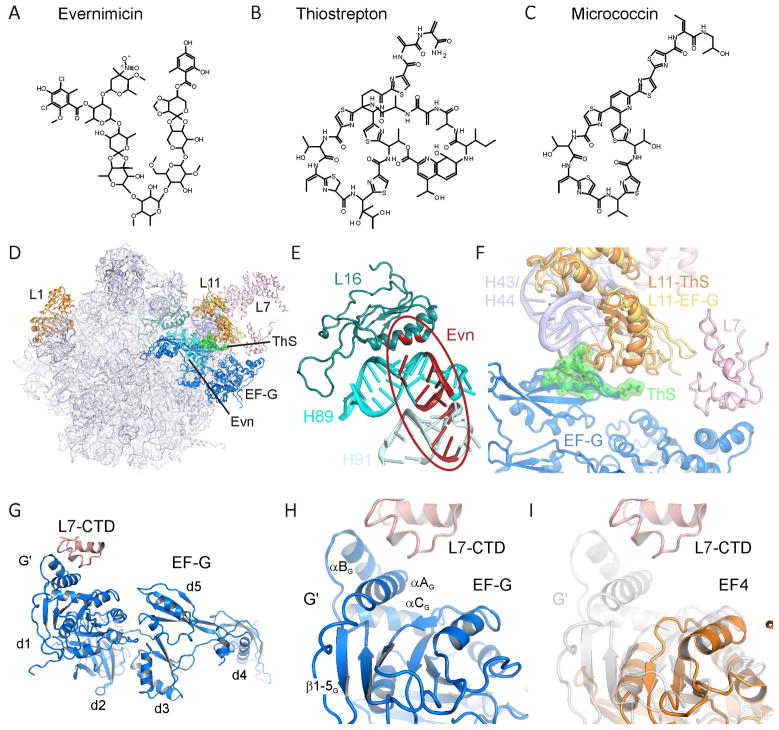
Figure 1. Chemical structures and ribosomal binding sites of thiopeptide and orthosomycin antibiotics.
Chemical structures of the (A) orthosomycin evernimicin (Evn), and the thiopeptide antibiotics (B) thiostrepton (ThS) and (C) micrococcin (MiC). (D) Overview of the binding sites of orthosomycins and thiopeptides on the large subunit relative to EF-G. R-proteins L1, L11 and L7 are shown for reference. (E) Putative binding site of orthosomycins spanning from H89 and H91 of the 23S rRNA. Residues highlighted in red have been associated biochemically with Evn or avilamycin (reviewed by Wilson, 2009). (F) Binding site of thiostrepton (green) in the cleft between H43 and H44 of the 23S rRNA and the N-terminal domain of L11 (L11-ThS) (Harms et al., 2008). The relative positions of EF-G (blue), C-terminal domain of L7/L12 (L7-CTD) and of a different conformation of L11 (L11-EF-G) are from (Gao et al., 2009). (G) Overview of domain arrangement of EF-G with contact between the L7-CTD and the G′ domain of EF-G as observed in the 70S-EF-G crystal structure (Gao et al., 2009). (H) Expansion of (G) highlighting the secondary structure elements of the G′ subdomain. (I) juxtaposition of the G′ subdomain of EF-G (grey transparency) with the G-domain of EF4 (orange) (Evans et al., 2008) that lacks a G′ subdomain. See also Figure S1.