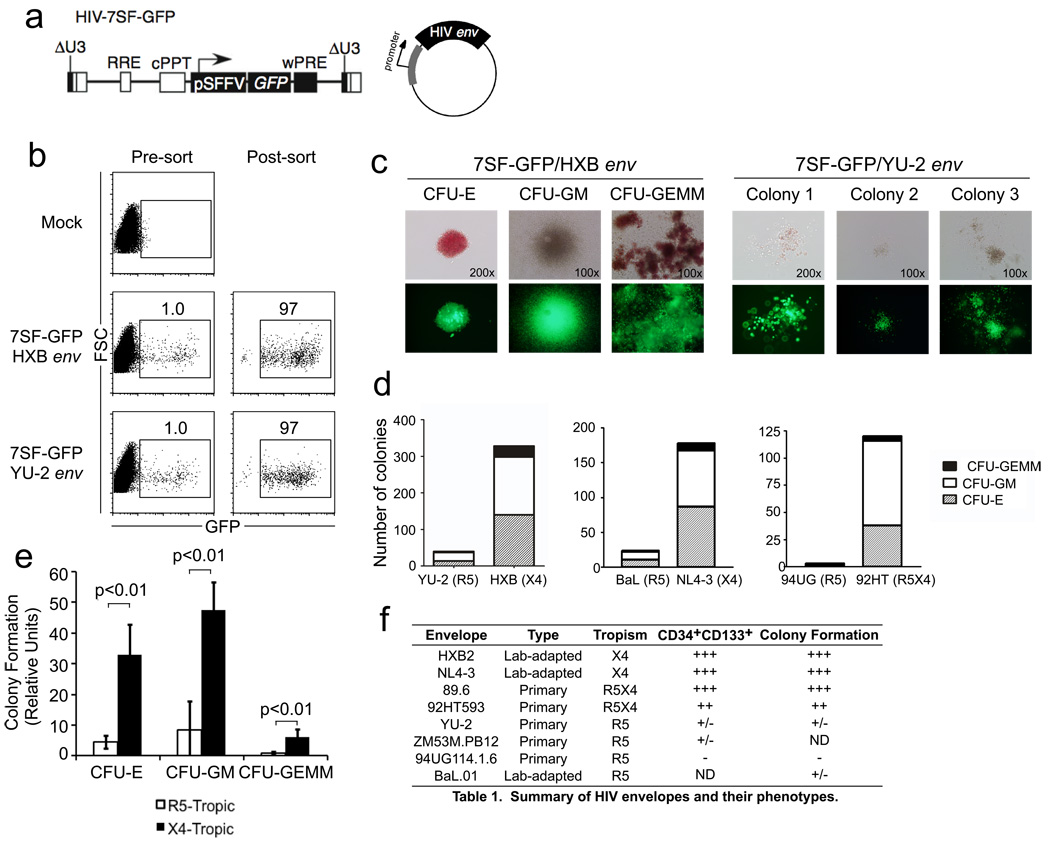
Figure 3. CXCR4-tropic HIV Envs infect HSPCs with the capacity to form multilineage colonies.
(a) Schematic of HIV-7SF-GFP construct and generic HIV envelope plasmid used to construct viruses used in b–e. (b) Flow cytometric analysis of cord-blood derived CD133+ cells infected with HIV-7SF-GFP pseudotyped with HXB (X4-tropic) or YU2 (R5-tropic) Env proteins, and purified by flow sorting. (c) Example colonies identified after culturing cells isolated as shown in part (b) for 14 days. Phase contrast and epifluorescence microscopy are shown (erythroid, CFU-E; myeloid, CFU-GM; or multilineage, CFU-GEMM). (d) Quantification of colony formation for the experiment shown in part (c) and for two similar experiments using other HIV Envs as indicated (erythroid, CFU-E; myeloid, CFU-GM; or multilineage, GFU-GEMM). The total number of colonies from 6 replicate wells is displayed. (e) Summary of colony formation results. Data were compiled from five independent experiments using six different Env proteins. The average normalized number of colonies observed with CXCR4- or dual-tropic Env versus CCR5-tropic Env is depicted. Error bars represent standard deviation and p-values were determined using the two-tailed Student’s T test (erythroid, CFU-E; myeloid, CFU-GM; or multilineage, GFU-GEMM). (f) Summary table of the ability of HIV-1 Envs of different tropism to infection CD133+CD34High and multipotent HSPCs.