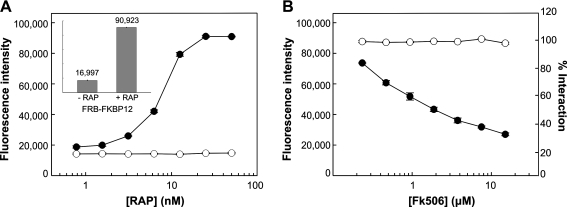
Fig. 2.
Chemically induced association and disruption of protein interactions with the M-PFC AB-TRIM assay. (A) Use of AB-TRIM assay to demonstrate (•) FRB-FKBP12 and (○) neg-FKBP12 (negative control) interaction in the presence RAP (0.78, 1.56, 3.13, 6.25, 12.5, 25.0, and 50.0 nM). Positive interaction of FRB-FKBP12 is shown by survival in TRIM 20 μg/mL media, as indicated by increasing fluorescence intensity of AB. Inset shows bar graph of FRB-FKBP12 interaction at 0 and 50 nM RAP, where relative fluorescence intensities are indicated. (B) Cells were pre-exposed to 12.5 nM RAP to facilitate FRB-FKBP12 interaction before treating them with FK506 (0.23, 0.47, 0.94, 1.88, 3.75, 7.50, and 15.0 μM). Step-wise inhibition of (•) FRB-FKBP12 interaction was observed with increasing FK506 concentration, where 100% interaction was defined as the fluorescence intensity at 0 μM FK506. Msm expressing the M-PFC construct (○) IdeR-IdeR was used as a viability and specificity control against FK506. Each data point represents the mean of triplicate wells ± SD. AB-TRIM, Alamar Blue–trimethoprim; Msm, Mycobacterium smegmatis; RAP, rapamycin; SD, standard deviation.