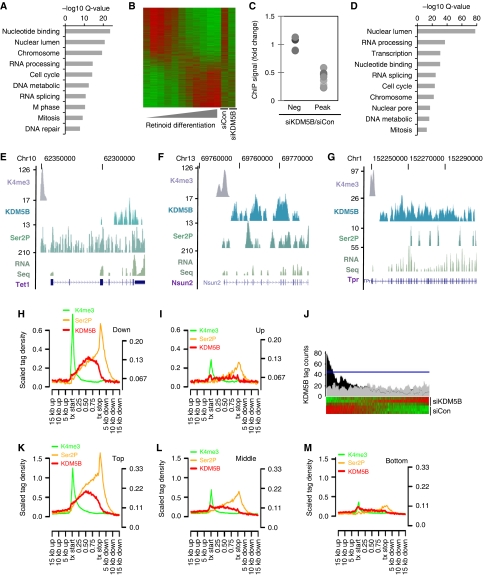
Figure 2.
An unexpected role for KDM5B in transcriptional activation. (A) Gene ontology (GO) analysis of microarray data showing categories of genes significantly down-regulated following siKDM5B. No up-regulated gene categories were detected at Q<1 × 10−6. (B) Heatmap comparison of microarray data from ESC retinoic acid (RA) differentiation and KDM5B knockdown experiments. (C) ChIP analysis of KDM5B ChIP-Seq peaks and negative control regions (Neg) following KDM5B knockdown (siKDM5B). Data are expressed as the ratio of siKDM5B ChIP signal over siCon. Each dot represents a distinct ChIP-Seq locus (pairwise comparisons to Peak data P<0.01). (D) GO analysis of RefSeq genes within 5 kb of a KDM5B ChIP-Seq peak 5′ end. (E–G) UCSC genome browser tracks depicting H3K4me3, KDM5B, and Ser 2 phosphorylated PolII (Ser2P) ChIP-Seq peaks at representative gene loci. The RNA-Seq track depicts 3′-directed RNA-Seq data from ESCs. The numbers on the left axes indicate peak amplitude. (H, I) ChIP-Seq tag density relative to significantly down- (H) and up-regulated (I) genes following KDM5B knockdown. All Ref-Seq gene lengths were scaled to 1. The left y axis corresponds to H3K4me3 ChIP-Seq data, while the right y axis represents other data. (J) Histogram of KDM5B ChIP-Seq peak tag density relative to ESC RefSeq microarray data rank ordered by fold-change following KDM5B knockdown. The black profile above the heat map depicts average number of significant ChIP-Seq KDM5B peak tags found per gene. The grey profile represents a randomized control and the blue line delineates an FDR of P<0.01. (K–M) ChIP-Seq tag density relative to the top (K), middle (L), and bottom third (M) of RefSeq genes expressed in ESCs. All Ref-Seq gene lengths were scaled to 1. The left y axis corresponds to H3K4me3 ChIP-Seq data, while the right y axis represents other data.