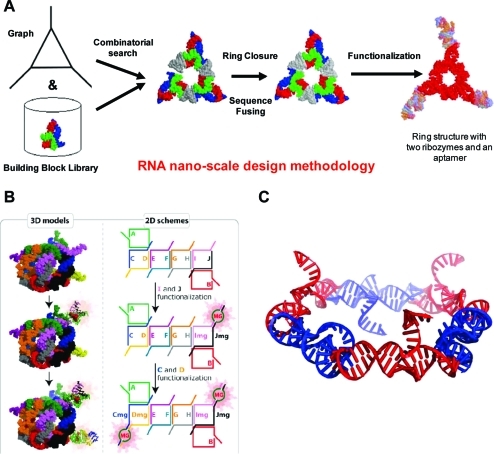
Figure 3.
(A) Representation of part of the computational RNA nanodesign pipeline associated with NanoTiler.63−65 What is depicted is one result, of many, which produces a closed-ring structure generated with the NanoTiler software that was derived in a totally automated fashion from a combinatorial search of motifs found in the RNAJunction database.(19) Sequences that were predicted for the shown structure were ultimately shown to be able to experimentally self-assemble into the depicted form. Panel A adapted with permission from ref (65). Copyright 2008 Elsevier. (B) Depiction of 10 stranded RNA cubes that were computer-designed with NanoTiler and later shown to be able to self-assemble.38,65 The layouts of the interacting strands are shown on the right. It should also be noted that the 10 stranded cubes, as shown here, were assembled in three forms: without a malachite green aptamer, with one malachite green aptamer, and with two malachite green aptamers. A properly formed aptamer fluoresces in the presence of a triarylmethane dye. The formation of the aptamers was used to confirm the cubes’ self-assembly and designed functionalization. Panel B adapted with permission from ref (38). Copyright 2011 Nature Publishing Group. (C) Depiction of the computer-designed, and later shown to be able to self-assemble, RNA hexagonal ring.34,68,69 Each corner of the ring contains the motif derived from the RNAIi/RNAIIi kissing loop interaction that forms an angle of about 120°, which is conducive to hexagonal ring formation. Sides and dangling ends can be further functionalized. Panel C adapted from ref (69). Copyright 2011 American Chemical Society.