Abstract
The cellulase activity of Bacillus subtilis AS3 was enhanced by optimizing the medium composition by statistical methods. The enzyme activity with unoptimised medium with carboxymethylcellulose (CMC) was 0.07 U/mL and that was significantly enhanced by CMC, peptone, and yeast extract using Placket-Burman design. The combined effects of these nutrients on cellulase activity were studied using 22 full factorial central composite design. The optimal levels of medium components determined were CMC (1.8%), peptone (0.8%), and yeast extract (0.479%). The maximum enzyme activity predicted by the model was 0.49 U/mL which was in good agreement with the experimental value 0.43 U/mL showing 6-fold increase as compared to unoptimised medium. The enzyme showed multisubstrate specificity, showing significantly higher activity with lichenan and β-glucan and lower activity with laminarin, hydroxyethylcellulose, and steam exploded bagasse. The optimised medium with lichenan or β-glucan showed 2.5- or 2.8-fold higher activity, respectively, at same concentration as of CMC.
1. Introduction
Cellulases (3.2.1.4) have a wide range of industrial applications such as textile, laundry, pulp and paper, fruit juice extraction, and animal feed additives as well as in bioethanol production [1]. The cellulases have great potential in saccharification of lignocellulosics to fermentable sugars which can be used for production of bioethanol, lactic acid, and single cell protein [2]. Majority of studies on cellulase production have focused on fungi, with relatively lesser emphasis on bacterial sources [1]. Cellulases from bacteria Bacillus, Clostridium, Cellulomonas, Thermomonospora, Ruminococcus, Bacteroides, Erwinia, Acetivibrio, and actinomycetes in particular Streptomyces species have been reported [3, 4]. Bacteria, due to their high natural diversity, faster growth have the capability to produce highly thermostable, alkali stable enzyme complement and may serve as highly potent sources of industrially important enzymes. Bacillus sp. continue to be dominant bacterial workhorses due to the capacity of some selected species to produce and secrete large quantities of extracellular enzymes [5–10]. Reports on strains belonging to species such as Bacillus sphaericus and Bacillus subtilis express high cellulose degradation activities [7, 10]. The production of extracellular cellulase in microorganisms is significantly influenced by a number of factors such as temperature, pH, aeration [11], and medium constituents [12]. The relationship between these variables has a marked effect on the ultimate production of the cellulase. There are reports on the influence of various fermentation parameters on cellulase production by different bacteria [11, 13] and fungi [14]. The traditional “one-variable-at-a-time approach” for medium optimisation disregards the complex interactions among various components. Statistically based experimental designs such as Placket-Burman design and response surface methodology (RSM) can be effectively used to study the effects of factors and searching optimum levels of parameters for desired responses.
In the present study the production of an alkaline extracellular cellulase by a novel strain of Bacillus subtilis (AS3) isolated from cow dung was enhanced by medium optimisation. The optimisation of the bioprocess was carried out through a stepwise experimental strategy including (1) screening the most significant factors affecting enzyme production using a two-level multifactorial Plackett-Burman design (2) optimisation of the most significant components and generating a mathematical model expressing the relationship between optimised factors and cellulase production by application of central composite design, and (3) verification of the model by monitoring the experimental production pattern. The effect of different carbon sources, lichenan and barley β-glucan, was also tested alternative to CMC in the basal culture media and optimised media to compare the enzyme production.
2. Materials and Methods
Carboxymethylcellulose (low viscosity, 50–200 cP), Lichenan and barley β-glucan was purchased from Sigma Aldrich (St Louis, USA). All other chemicals used in the study were of highest purity grade commercially available.
2.1. Microorganism and Cultivation Conditions
The organism used for the study is Bacillus subtilis AS3 (Genebank accession No. EU754025). The culture was maintained in nutrient agar slant with filter paper strip at 4°C and subcultured every 2 weeks. A loopful of culture from the nutrient agar slant was transferred to 5 mL of nutrient broth. The culture was grown at 37°C with 180 rpm for 24 h. 2% of the culture inoculum was transferred to 250 mL Erlenmeyer flask containing 50 mL of cellulolytic medium described by [15] with modification to contain (g/L): CMC, 10; peptone, 5; yeast extract, 5; K2HPO4, 1; MgSO4·7H2O, 0.25; FeSO4·7H2O, 0.25; MnCl2·4H2O, 0.5. The pH of the medium was adjusted to 7.0 using 1 N NaOH before autoclaving. All the experiments were performed in duplicate. The culture broth after 36 h of incubation was centrifuged at 10,000 g for 10 min at 4°C to separate the cells. The cell-free supernatant was analyzed for enzyme activity and protein concentration.
2.2. Assay of Enzyme Activity
The assay of cellulase was carried out in 100 μL enzyme-carboxymethylcellulose reaction mixture containing 65 μL of 2% CMC (final concentration, 1.3%) in 50 mM Glycine NaOH buffer pH 9.2 and 35 μL of cell-free supernatant and incubated at 45°C for 10 min. The assay conditions for the enzyme from the new isolate of Bacillus subtilis AS3 were optimised (data not shown). The enzyme had an optimum pH of 9.2 so it was called alkaline cellulase and showed maximum activity at 45°C. The cellulase activity was calculated by estimation of reducing sugars liberated from carboxymethylcellulose (CMC). The reducing sugar was estimated by the method of Nelson and Somogyi [16, 17]. A standard curve was prepared with glucose. One unit (U) of cellulase activity is defined as the amount of enzyme that liberates 1 μmole of reducing sugar (glucose) per min at 45°C in 50 mM Glycine NaOH buffer, pH 9.2. The protein concentration was determined by the Bradford method using bovine serum albumin (BSA) as standard [18].
2.3. Screening of the Most Significant Medium Components by Plackett-Burman Design
For the selection of significant medium, components for cellulase production such as carbon source, nitrogen source, and inorganic salts were screened and identified by the Plackett-Burman design using statistical software package MINITAB (Release 15.1, PA, USA). A total of seven parameters CMC, yeast extract, peptone, K2HPO4, MnCl2·4H2O, MgSO4·7H2O, and FeSO4·7H2O were considered for screening with each factor examined in two levels: −1 for low level and +1 for high level [19]. Plackett-Burman experimental design is based on the first-order polynomial model:
| (1) |
where Y is the response (enzyme activity), β0 is the model intercept, βi is the linear coefficient, and Xi is the level of the independent variable. This model does not describe interaction among factors and it is used to screen and evaluate the important factors that influence the response. All experiments were carried out in duplicate and the averages of the cellulase activity were taken as the response (Table 1). From the regression analysis the variables, which were significant at or above 95% level (P < .05), were considered to have greater impact on cellulase activity and were further optimised by central composite design.
Table 1.
Plackett-Burman design in coded units and real values (in parenthesis) in g/L for seven variables along with the cellulase activity.
| Run Order |
CMC (X1) | Peptone (X2) | YE (X3) | K2HPO4 (X4) | MgSO4·7H2O (X5) | FeSO4·7H2O (X6) | MnCl2·4H2O (X7) | E.A (U/mL) |
|---|---|---|---|---|---|---|---|---|
| (1) | +1 (18) | −1 (2) | −1 (1) | −1 (0.5) | −1 (0.05) | +1 (0.45) | −1 (0.01) | 0.084 |
| (2) | −1 (2) | −1 (2) | −1 (1) | −1 (0.5) | +1 (0.45) | −1 (0.05) | +1 (0.1) | 0.003 |
| (3) | −1 (2) | +1 (8) | +1 (9) | +1 (2) | +1 (0.45) | −1 (0.05) | −1 (0.01) | 0.080 |
| (4) | +1 (18) | −1 (2) | +1 (9) | +1 (2) | −1 (0.05) | −1 (0.05) | −1 (0.01) | 0.226 |
| (5) | −1 (2) | −1 (2) | −1 (1) | +1 (2) | −1 (0.05) | +1 (0.45) | −1 (0.01) | 0.004 |
| (6) | +1 (18) | +1 (8) | +1 (9) | +1 (2) | −1 (0.05) | −1 (0.05) | +1 (0.1) | 0.252 |
| (7) | +1 (18) | +1 (8) | −1 (1) | +1 (2) | +1 (0.45) | −1 (0.05) | −1 (0.01) | 0.221 |
| (8) | −1 (2) | +1 (8) | +1 (9) | −1 (0.5) | +1 (0.45) | +1 (0.45) | −1 (0.01) | 0.026 |
| (9) | +1 (18) | −1 (2) | +1 (9) | +1 (2) | +1 (0.45) | +1 (0.45) | −1 (0.01) | 0.205 |
| (10) | −1 (2) | −1 (2) | −1 (1) | −1 (0.5) | −1 (0.05) | −1 (0.05) | −1 (0.01) | 0.006 |
| (11) | −1 (2) | +1 (8) | +1 (9) | −1 (0.5) | −1 (0.05) | −1 (0.05) | −1 (0.01) | 0.049 |
| (12) | +1 (18) | +1 (8) | −1 (1) | −1 (0.5) | +1 (0.45) | +1 (0.45) | −1 (0.01) | 0.168 |
| (13) | +1 (18) | −1 (2) | −1 (1) | +1 (2) | +1 (0.45) | −1 (0.05) | +1 (0.1) | 0.102 |
| (14) | +1 (18) | −1 (2) | +1 (9) | −1 (0.5) | +1 (0.45) | +1 (0.45) | +1 (0.1) | 0.197 |
| (15) | −1 (2) | −1 (2) | +1 (9) | −1 (0.5) | +1 (0.45) | −1 (0.05) | +1 (0.1) | 0.024 |
| (16) | −1 (2) | +1 (8) | −1 (1) | +1 (2) | +1 (0.45) | +1 (0.45) | +1 (0.1) | 0.003 |
| (17) | +1 (18) | +1 (8) | −1 (1) | −1 (0.5) | −1 (0.05) | −1 (0.05) | +1 (0.1) | 0.214 |
| (18) | +1 (18) | +1 (8) | +1 (9) | −1 (0.5) | −1 (0.05) | +1 (0.45) | +1 (0.1) | 0.249 |
| (19) | −1 (2) | +1 (8) | −1 (1) | +1 (2) | −1 (0.05) | +1 (0.45) | +1 (0.1) | 0.025 |
| (20) | −1 (2) | −1 (2) | +1 (9) | +1 (2) | −1 (0.05) | +1 (0.45) | +1 (0.1) | 0.022 |
2.4. Central Composite Design (CCD) and Statistical Analysis
A 22 full-factorial central composite design (CCD) with three medium constituents, that is, CMC, peptone, and yeast extract at five coded levels, was generated by Minitab statistical software (Release 15). In this study, the experimental plan consisted of 20 runs ( = 2k + 2k + n0), where “k” is the number of independent variables and n0 the number of replicate runs at center point of the variables. The relationships and interrelationships of the variables were determined by fitting the second-order polynomial equation to data obtained from 20 experiments
| (2) |
where Y is the predicted response, k is the number of factor variables, β0 is the model constant, βi is the linear coefficient, βii is the quadratic coefficient, and βij is the interaction coefficient. The following equation was used for coding the actual experimental values of the factors in the range of (−1 to +1):
| (3) |
where xi is the dimensionless value of an independent variable, Xi is the real value of an independent variable, X0 is the value of Xi at the center point, and ΔXi is the step change. Statistical analysis of the data was performed by design package Design Expert 7.0 to evaluate the analysis of variance (ANOVA) to determine the significance of each term in the equations fitted and to estimate the goodness of fit in each case. The fitted polynomial equation was then expressed in the form of three-dimensional response surface plots to illustrate the main and interactive effects of the independent variables on the dependent ones. The combination of different optimised variables, which yielded the maximum response, was determined to verify the validity of the model. In order to verify the accuracy of the predicted model an experiment was conducted with initial and optimised media.
2.5. Effect of Different Cellulosic Substrates on Cellulase Production
The culture was grown under same conditions except replacing 1% CMC with 1% Lichenan or 1% barley β-glucan as carbon source in unoptimised and replacing 1.8% CMC with 1.8% lichenan or 1.8% barley β-glucan in the optimised medium. The enzyme production was studied at different time intervals of 12 h, 24 h, 36 h, 48 h, and 60 h. The enzyme assay was performed with final concentration of 1% lichenan or 1% barley β-glucan as substrate in 50 mM Glycine NaOH buffer (pH 9.2) incubated at 45°C for 10 min. The cellulase activity was calculated as described earlier.
3. Results
3.1. Screening of the Most Significant Medium Components by Plackett-Burman Design
The cellulase activity of the cell-free supernatant of Bacillus subtilis AS3 culture at 36 h was maximum. The data in Table 1 indicated that there was a wide variation of cellulase activity from 0.003 U/mL to 0.25 U/mL in twenty trials. This variation reflected the significance of factors on the enzyme activity. The analysis of regression coefficients and t-value of seven ingredients are shown in Table 2. Generally, a large t-value associated with a low P-value of a variable indicates a high significance of the corresponding model term. CMC, peptone, yeast extract, K2HPO4, and MnCl2·4H2O displayed a positive effect for enzyme production, whereas MgSO4·7H2O and FeSO4·7H2O had a negative effect on enzyme activity (Table 2). The variables with confidence levels greater than 95% were considered as significant. CMC was significant at 100% confidence levels for cellulase production, and peptone and yeast extract were found significant at 99 and 99.7% levels, respectively, for cellulase activity. None of the components had significant negative effect. Neglecting the variables which were insignificant, the model equation for cellulase activity can be written as
Table 2.
Statistical analysis of Plackett Burman design showing coefficient values, t- and P-value for each variable.
| Variable | Coefficient | t-Stat | P-value | Confidence level (%) |
|---|---|---|---|---|
| Intercept | 0.107817 | 15.96 | .000 | 100 |
| CMC (X1) | 0.083852 | 12.41 | .000 | 100 |
| Peptone (X2) | 0.020703 | 3.06 | .01 | 99 |
| Yeast Extract (X3) | 0.024881 | 3.68 | .003 | 99.7 |
| K2HPO4 (X4) | 0.005974 | 0.88 | .394 | 60.6* |
| MgSO4·7H2O (X5) | −0.005107 | −0.76 | .464 | 53.6* |
| FeSO4·7H2O (X6) | −0.009733 | −1.44 | .175 | 82.5* |
| MnCl2·4H2O (X7) | 0.001163 | 0.17 | .866 | 13.4* |
*Insignificant values.
| (4) |
where X1 = CMC, X2 = peptone, X3 = yeast extract. The nutrient CMC showed positive effect. The +1 level of CMC was chosen at 1.8% as higher concentration would cause increased viscosity, making the separation of cells difficult. Different nitrogen sources, such as yeast extract and peptone were chosen, as these have been reported to significantly affect the cellulase production [20]. On the basis of the calculated t-values (Table 2) CMC, peptone, and yeast extract were chosen for further optimisation, since these factors had the most significant effect on the cellulase activity. All other variables used in all the trials were kept to the median level.
3.2. Optimisation of Medium Components by CCD
At the end of screening experiments by Plackett-Burman design three factors were found to play a significant role in cellulase production. The respective low and high levels of each variable with the coded levels in parenthesis along with the CCD design with response (U/mL) are given in Table 3. The results of the second-order response surface model fitting in the form of ANOVA are given in Table 4. To test the fit of the model equation, the regression-based determination coefficient R2 was evaluated. The nearer the values of R2 to 1, the model would explain better for variability of experimental values to the predicted values [21]. The model presented a high determination coefficient (R2 = 0.9911) explaining 99% of the variability in the response (Table 4). The coefficients of regression were calculated and the following regression equation was obtained:
Table 3.
Full factorial central composite design matrix of three variables in coded units and real values (in parenthesis) with experimental and predicted response.
| Run No | CMC (X1) (g/L) | Peptone (X2) (g/L) | Yeast extract (X3) (g/L) | Enzyme activity (U/mL) | |
|---|---|---|---|---|---|
| Predicted | Observed | ||||
| (1) | 0 (10) | 0 (5) | 0 (5) | 0.270 | 0.271 |
| (2) | 0 (10) | 0 (5) | 0 (5) | 0.270 | 0.271 |
| (3) | 1 (14.76) | −1 (3.22) | 1 (7.38) | 0.341 | 0.330 |
| (4) | −1 (5.24) | −1 (3.22) | −1 (2.62) | 0.122 | 0.123 |
| (5) | 0 (10) | 0 (5) | 0 (5) | 0.270 | 0.272 |
| (6) | 0 (10) | 0 (5) | 0 (5) | 0.270 | 0.261 |
| (7) | − α (2) | 0 (5) | 0 (5) | 0.118 | 0.119 |
| (8) | 1 (14.76) | −1 (3.22) | −1 (2.62) | 0.313 | 0.306 |
| (9) | 0 (10) | 0 (5) | 0 (5) | 0.270 | 0.274 |
| (10) | −1 (5.24) | −1 (3.22) | 1 (7.38) | 0.221 | 0.208 |
| (11) | −1 (5.24) | 1 (3.22) | 1 (7.38) | 0.215 | 0.214 |
| (12) | 0 (10) | 0 (5) | 0 (5) | 0.270 | 0.271 |
| (13) | + α (18) | 0 (5) | 0 (5) | 0.422 | 0.431 |
| (14) | 1 (14.76) | 1 (6.78) | 1 (7.38) | 0.385 | 0.377 |
| (15) | 1 (14.76) | 1 (6.78) | −1 (2.62) | 0.364 | 0.371 |
| (16) | 0 (10) | − α (2.0) | 0 (5) | 0.269 | 0.283 |
| (17) | −1 (5.24) | 1 (6.78) | −1 (2.62) | 0.123 | 0.128 |
| (18) | 0 (10) | 0 (5) | +α (9.0) | 0.276 | 0.292 |
| (19) | 0 (10) | +α (8.0) | 0 (5) | 0.307 | 0.302 |
| (20) | 0 (10) | 0 (5) | − α (1.0) | 0.175 | 0.168 |
Table 4.
ANOVA for quadratic model.
| Source | SS | DF | MS | F-value | Prob (P) > F |
|---|---|---|---|---|---|
| Model | 0.133841 | 9 | 0.014871 | 123.94 | .000 |
| Residual (error) | 0.001200 | 10 | 0.000120 | ||
| Lack of fit | 0.001087 | 5 | 0.000217 | 9.66 | .013 |
| Pure error | 0.000113 | 5 | 0.000023 | ||
| Total | 0.135040 | 19 | |||
R 2 = 0.9911; Adj R2 = 0.9831
SS, sum of squares; DF, Degree of freedom; MS, mean square.
| (5) |
where Y = response (cellulase activity), X1 = CMC, X2 = peptone, X3 = yeast extract in coded values. The statistical significance of (5) was checked by F test, the results of ANOVA are shown in Table 4. The results demonstrated that the model is highly significant and is evident from Fischer's F-test with a very low probability value (P model > F =.0000) (Table 4). Model coefficients estimated by regression analysis for each variable is shown in Table 5. The significance of each coefficient was determined by t-values and P-values. The larger the magnitude of t-test value and smaller the P-value indicates the high significance of the corresponding coefficient [21]. The results revealed that CMC concentration (X1) had a significant effect (P < .0000) on cellulase production than the effects of other variables. The P-values (<.0001) and lack of fit (.013) for the model suggested that the obtained experimental data were in good fit. Three-dimensional response surface plots were constructed by plotting the response (enzyme activity) on the Z-axis against any two independent variables, while maintaining other variables at their median levels shown in Figures 1(a)–1(c).
Table 5.
Model coefficient estimated by multiple linear regressions.
| Model term | Parameter estimate | Standard error | Computed t-value | P-value |
|---|---|---|---|---|
| Intercept | 0.270187 | 0.004467 | 60.479 | .000 |
| X1 (CMC) | 0.151937 | 0.004985 | 30.480 | .000 |
| X2 (Peptone) | 0.019066 | 0.004985 | 3.825 | .003 |
| X3 (Yeast extract) | 0.050437 | 0.004985 | 10.118 | .000 |
| X12 | 0.000199 | 0.008161 | 0.024 | .981 |
| X22 | 0.017596 | 0.008161 | 2.16 | .056 |
| X32 | −0.045086 | 0.008161 | −5.524 | .000 |
| X1∗X2 | 0.035305 | 0.010954 | 3.223 | .009 |
| X1∗X3 | −0.049972 | 0.010954 | −4.562 | .001 |
| X2∗X3 | −0.005799 | 0.010954 | −0.529 | .608 |
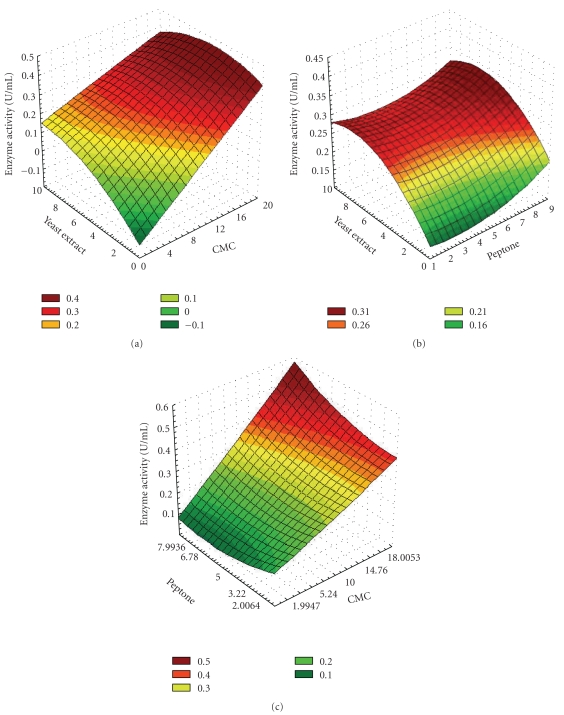
Figure 1.
Three-dimensional response surface plot for alkaline cellulase production showing the interactive effects of medium components in g/L: (a) yeast extract and CMC median level: peptone, 5 g/L, (b) yeast extract and peptone, median level: CMC, 10 g/L; (c) CMC and peptone, median level: yeast extract, 5 g/L.
The response surfaces having circular contour plot indicate no interaction, whereas, an elliptical or saddle nature of the contour plot indicates significant interaction between the corresponding variables. Figure 1(a) shows that with increase in CMC concentration in the entire range, surface plot is sharply ascending indicating an enhancement of enzyme activity. Similar ascending nature of surface plot was observed with yeast extract however beyond 5 g/L of yeast extract the surface tends to decline indicating a decrease in enzyme activity. This proves a strong interaction between CMC and yeast extract. This is also confirmed by Student's t-test with P-value (P = .001) and t-value (−4.562) as shown in Table 5. Figure 1(b) shows that, with the increase in yeast extract concentration to mid range, the surface is ascending indicating enhancement of enzyme activity and beyond the midrange the surface curvature is declining. But no significant change in surface curvature is observed with increase in peptone concentration showing an insignificant interaction. Figure 1(c) shows that, with the increase in CMC concentration, the surface is ascending indicating enhancement of enzyme activity, and also, in case of peptone the surface is ascending showing a significant interaction. This interaction is also confirmed by Student's t-test with P-value (P = .009) and t-value (3.223) as shown in Table 5. Therefore in both Figures 1(a) and 1(c) there was a steep increase in cellulase activity with increase in CMC concentration up to the maximum level of 18 g/L. The optimum levels of the variables were obtained by solving the regression equation and also by analyzing the response surface contour plots using Design Expert software. The model predicted a maximum cellulase activity of 0.49 U/mL appearing at CMC (18 g/L), peptone (8 g/L), and yeast extract (4.798 g/L) by keeping the other components at their median levels.
3.3. Experimental Validation of the Model
To validate the predicted model an experiment was conducted in duplicate using the optimum medium composition. Cellulase activity of 0.43 U/mL was observed at this optimised medium composition. Under these optimised conditions, the predicted response for alkaline cellulase production was determined to be 0.49 U/mL, and the observed experimental value was found to be 0.43 U/mL. The excellent correlation between predicted and experimental values of this experiment justifies the validity of the response model and the existence of an optimum point.
3.4. Effect of B-Glucan and Lichenan on Enzyme Activity by Replacing CMC in Optimised Medium
The alkaline cellulase from Bacillus subtilis AS3 showed multisubstrate specificity showing activity with CMC, laminarin, hydroxyethylcellulose, and steam exploded bagasse and significantly higher activity with lichenan and barley β-glucan. The effect of cellulosic substrates with different linkages on enzyme production was studied as an alternative to CMC. It was observed that with lichenan or barley β-glucan as carbon source the activity reached maximum at 12 h and remained stable till 36 h (Table 6). The optimised medium showed 3.4- and 3.5-fold increase in activity with lichenan (1.2 U/mL) and barley β-glucan (1.4 U/mL), respectively, compared to the unoptimised medium (0.35 U/mL) and 0.4 (U/mL) (Table 6). On comparison of the activity with CMC optimised medium (0.43 U/mL) the increase was 2.8- or 3.2-fold higher with the optimised medium containing lichenan or barley β-glucan, respectively (Table 6). The enzyme can be called β-1,3-1,4-glucanase (lichenase) which cleaves the β-1,3-1,4 mixed linkages.
Table 6.
Variation of enzyme activity with CMC, lichenan, and barley β-glucan with and without medium optimisation.
| Media | CMC | Lichenan | β-Glucan |
|---|---|---|---|
| Unoptimised (U/mL) | 0.07 | 0.35 | 0.4 |
| Optimised (U/mL) | 0.43 | 1.2 | 1.4 |
| Fold increase | 6 | 3.4 | 3.5 |
| Maximum activity (h) | 36 | 12 | 12 |
4. Discussion
Considering the commercial importance of alkaline cellulase, we have attempted to optimise the medium components for its enhanced production from Bacillus subtilis (AS3). The significant variables for enhancing alkaline cellulase production were screened and selected using the Plackett-Burman design. The results showed that CMC, peptone, and yeast extract significantly and positively affected the enzyme production from Bacillus subtilis (AS3). It has been reported that CMC shows inducing effect on cellulase production. Ahamed and Vermette and Domingues et al. reported that biosynthesis of cellulases in Trichoderma reesei was very high in medium with carboxymethylcellulose as carbon source [22, 23]. It was also reported that yeast extract and peptone have significant effect on cellulase production [20]. These three ingredients were used as variables in the response surface analysis. Under the optimal medium composition (CMC, 18 g/L; peptone, 8 g/L and yeast extract, 4.79 g/L) the experimental value of enzyme activity 0.43 U/mL perfectly matched with predicted value of 0.49 U/mL showing 6-fold increase with respect to the control basal medium which showed only 0.07 U/mL enzyme activity. These results propose a medium formulation that could serve as a basal medium for further optimisation studies. The optimised enzyme activity value obtained in this study is much higher than the reported values with other strains. Li et al. reported maximum cellulase activity (0.26 U/mL) of a Bacillus sp. when the culture was grown in LB medium supplemented with 1% CMC [20]. It has been reported recently that Bacillus sp. (DUSELR13) and Brevibacillus sp. (DUSELG12) isolated from gold mine produced maximum CMCase activity 0.12 U/mL and 0.02 U/mL, respectively, under unoptimised conditions [8]. In Geobacillus sp. it has been reported that optimizing the culture conditions and factorial designs and additions of yeast extract and ammonium sulfate resulted in twofold increase in cellulase production from 0.425 U/mL (basal medium) to 0.8 U/mL under optimised conditions [24]. In another study a much lower cellulase activity of 0.0113 U/mL was observed under optimised conditions from Geobacillus sp. [25]. Rastogi et al. reported maximum CMCase and cellulase activities of 0.058 and 0.043 U/mL, respectively, from cell-free culture supernatants of Geobacillus sp. isolated from deep goldmine environment [26]. Ariffin et al. recorded maximum FPase, CMCase, and β-glucosidase activities of 0.011, 0.079, and 0.038 U/mL, respectively, by Bacillus pumilus EB3 produced in a 2L stirred tank reactor [6]. Liang et al. reported basal level cellulase activity of FPU 0.02 U/mL in the crude culture supernatant of Brevibacillus sp. JXL [27]. Moreover, Bacillus subtilis CK-2 isolated from compost soil and Bacillus megaterium were reported producing CMCase activity of 0.26 U/mL and 0.102 U/mL, respectively, with 1% CMC [5, 28]. Although the cellulase activity is lower when compared to the activity of fungal strains but the fact can be taken into account that the strain Bacillus subtilis AS3 (EU 754028) has high β-glucanase and lichenase activity (that can break β-1,3-1,4 linkages) as compared to carboxymethylcellulase activity. It was observed that in comparison to optimised medium with CMC as substrate it showed 2.8- and 3-fold increase with lichenan and β-glucan, respectively, and the activity reached maximum at 12 h in contrary to CMC substrate which requires 36 h to reach maximum activity. Therefore the isolate is a high β-1,3-1,4-glucanase (lichenase) producing strain. Considering this property the enzyme isolated from this novel strain (Bacillus subtilis AS3 EU 754025) which is alkaline in nature, thermostable and active over a wide range of pH can be used for various applications.
5. Conclusion
Medium components for higher cellulase activity from Bacillus subtilis AS3 were optimised by RSM. Using Placket-Burman design CMC, peptone and yeast extract significantly enhanced enzyme activity. A 22 full factorial central composite design was applied to study the combined effects of these nutrients. The optimal levels of components were CMC (1.8%), peptone (0.8%), and yeast extract (0.479%). The model predicted maximum cellulase activity of 0.49 U/mL which was in perfect agreement with experimental value of 0.43 U/mL, showing 6-fold increase as compared to unoptimised medium that gave an activity of 0.07 U/mL. The enzyme showed significantly higher activity with lichenan and β-glucan. The Bacillus subtilis AS3 strain is being used for degradation of cellulosic substrates such as rice husk, sugarcane bagasse, and wild grass.
Acknowledgment
The research work was supported by a joint project grant from Department of Biotechnology, Ministry of Science and Technology, New Delhi, India, to A. Goyal and D. Goyal.
References
- 1.Bhat MK. Cellulases and related enzymes in biotechnology. Biotechnology Advances. 2000;18(5):355–383. doi: 10.1016/s0734-9750(00)00041-0. [DOI] [PubMed] [Google Scholar]
- 2.Maki M, Leung KT, Qin W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. International Journal of Biological Sciences. 2009;5(5):500–516. doi: 10.7150/ijbs.5.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robson LM, Chambliss GH. Cellulases of bacterial origin. Enzyme and Microbial Technology. 1989;11(10):626–644. [Google Scholar]
- 4.Nascimento RP, Junior NA, Pereira N, Jr., Bon EPS, Coelho RRR. Brewer’s spent grain and corn steep liquor as substrates for cellulolytic enzymes production by Streptomyces malaysiensis. Letters in Applied Microbiology. 2009;48(5):529–535. doi: 10.1111/j.1472-765X.2009.02575.x. [DOI] [PubMed] [Google Scholar]
- 5.Aa K, Flengsrud R, Lindahl V, Tronsmo A. Characterization of production and enzyme properties of an endo-β-1 ,4-glucanase from Bacillus subtilis CK-2 isolated from compost soil. Antonie Van Leeuwenhoek. 1994;66(4):319–326. doi: 10.1007/BF00882767. [DOI] [PubMed] [Google Scholar]
- 6.Ariffin H, Abdullah N, Umi Kalsom MS, Shirai Y, Hassan MA. Production and characterization of cellulase by Bacillus pumilus EB3. International Journal of Engineering and Technology. 2006;3:47–53. [Google Scholar]
- 7.Mawadza C, Boogerd FC, Zvauya R, Van Verseveld HW. Influence of environmental factors on endo-β-1,4-glucanase production by Bacillus HR 68, isolated from a Zimbabwean hot spring. Antonie Van Leeuwenhoek. 1996;69(4):363–369. doi: 10.1007/BF00399625. [DOI] [PubMed] [Google Scholar]
- 8.Rastogi G, Bhalla A, Adhikari A, et al. Characterization of thermostable cellulases produced by Bacillus and Geobacillus strains. Bioresource Technology. 2010;101(22):8798–8806. doi: 10.1016/j.biortech.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Schallmey M, Singh A, Ward OP. Developments in the use of Bacillus species for industrial production. Canadian Journal of Microbiology. 2004;50(1):1–17. doi: 10.1139/w03-076. [DOI] [PubMed] [Google Scholar]
- 10.Singh J, Batra N, Sobti RC. Purification and characterisation of alkaline cellulase produced by a novel isolate Bacillus sphaericus JS1. Journal of Industrial Microbiology and Biotechnology. 2004;31(2):51–56. doi: 10.1007/s10295-004-0114-0. [DOI] [PubMed] [Google Scholar]
- 11.Immanuel G, Dhanusha R, Prema P, Palavesam A. Effect of different growth parameters on endoglucanase enzyme activity by bacteria isolated from coir retting effluents of estuarine environment. International Journal of Environmental Science and Technology. 2006;3(1):25–34. [Google Scholar]
- 12.Prasertsan P, Doelle HW. Nutrient optimization for cellulase biosynthesis by a newly isolated Cellulomonas sp. World Journal of Applied Microbiology and Biotechnology. 1987;3(1):33–43. [Google Scholar]
- 13.Rajoka MI. Influence of various fermentation variables on exo-glucanase production in Cellulomonas flavigena. Electronic Journal of Biotechnology. 2004;7(3):256–263. [Google Scholar]
- 14.Stewart JC, Parry JB. Factors influencing the production of cellulase by Aspergillus fumigatus (Fresenius) Journal of General Microbiology. 1981;125(1):33–39. doi: 10.1099/00221287-125-1-33. [DOI] [PubMed] [Google Scholar]
- 15.Horikoshi K, Nakao M, Kurono Y, Sashihara N. Cellulases of an alkalophilic Bacillus strain isolated from soil. Canadian Journal of Microbiology. 1984;30(6):774–779. [Google Scholar]
- 16.Nelson N. A photometric adaptation of the Somogyi method for the determination of glucose. Journal of Biological Chemistry. 1944;153:375–380. [Google Scholar]
- 17.Somogyi M. A new reagent for the determination of sugars. Journal of Biological Chemistry. 1945;160:61–68. [Google Scholar]
- 18.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika. 1946;33:305–325. [Google Scholar]
- 20.Li W, Zhang WW, Yang MM, Chen YL. Cloning of the thermostable cellulase gene from newly isolated Bacillus subtilis and its expression in Escherichia coli. Molecular Biotechnology. 2008;40(2):195–201. doi: 10.1007/s12033-008-9079-y. [DOI] [PubMed] [Google Scholar]
- 21.Purama RK, Goyal A. Screening and optimization of nutritional factors for higher dextransucrase production by Leuconostoc mesenteroides NRRL B-640 using statistical approach. Bioresource Technology. 2008;99(15):7108–7114. doi: 10.1016/j.biortech.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 22.Ahamed A, Vermette P. Culture-based strategies to enhance cellulase enzyme production from Trichoderma reesei RUT-C30 in bioreactor culture conditions. Biochemical Engineering Journal. 2008;40(3):399–407. [Google Scholar]
- 23.Domingues FC, Queiroz JA, Cabral JMS, Fonseca LP. The influence of culture conditions on mycelial structure and cellulase production by Trichoderma reesei Rut C-30. Enzyme and Microbial Technology. 2000;26(5-6):394–401. doi: 10.1016/s0141-0229(99)00166-0. [DOI] [PubMed] [Google Scholar]
- 24.Abdel-Fattah YR, El-Helow ER, Ghanem KM, Lotfy WA. Application of factorial designs for optimization of avicelase production by a thermophilic Geobacillus isolate. Research Journal of Microbiology. 2007;2(1):13–23. [Google Scholar]
- 25.Tai SK, Lin HPP, Kuo J, Liu JK. Isolation and characterization of a cellulolytic Geobacillus thermoleovorans T4 strain from sugar refinery wastewater. Extremophiles. 2004;8(5):345–349. doi: 10.1007/s00792-004-0395-2. [DOI] [PubMed] [Google Scholar]
- 26.Rastogi G, Muppidi GL, Gurram RN, et al. Isolation and characterization of cellulose-degrading bacteria from the deep subsurface of the Homestake gold mine, Lead, South Dakota, USA. Journal of Industrial Microbiology and Biotechnology. 2009;36(4):585–598. doi: 10.1007/s10295-009-0528-9. [DOI] [PubMed] [Google Scholar]
- 27.Liang Y, Yesuf J, Schmitt S, Bender K, Bozzola J. Study of cellulases from a newly isolated thermophilic and cellulolytic Brevibacillus sp. strain JXL. Journal of Industrial Microbiology and Biotechnology. 2009;36(7):961–970. doi: 10.1007/s10295-009-0575-2. [DOI] [PubMed] [Google Scholar]
- 28.Beukes N, Pletschke BI. Effect of sulfur-containing compounds on Bacillus cellulosome-associated ‘CMCase’ and ‘Avicelase’ activities. FEMS Microbiology Letters. 2006;264(2):226–231. doi: 10.1111/j.1574-6968.2006.00465.x. [DOI] [PubMed] [Google Scholar]