Abstract
Anurans (frogs and toads) are unique among land vertebrates in possessing a free-living larval stage that, parallel to adult frogs, diversified into an impressive range of ecomorphs. The tempo and mode at which tadpole morphology evolved through anuran history as well as its relationship to lineage diversification remain elusive. We used a molecular phylogenetic framework to examine patterns of morphological evolution in tadpoles in light of observed episodes of accelerated lineage diversification. Our reconstructions show that the expansion of tadpole morphospace during the basal anuran radiation in the Triassic/Early Jurassic was unparalleled by the basal neobatrachian radiation in the Late Jurassic/Early Cretaceous or any subsequent radiation in the Late Cretaceous/Early Tertiary. Comparative analyses of radiation episodes indicate that the slowdown of morphospace expansion was caused not only by a drop in evolutionary rate after the basal anuran radiation but also by an overall increase in homoplasy in the characters that did evolve during later radiations. The overlapping sets of evolving characters among more recent radiations may have enhanced tadpole diversity by creating unique combinations of homoplastic traits, but the lack of innovative character changes prevented the exploration of fundamental regions in morphospace. These complex patterns transcend the four traditionally recognized tadpole morphotypes and apply to most tissue types and body parts.
A key feature that distinguishes anurans from any other living land vertebrate is a biphasic life history in which larval (tadpole) and adult (frog) stages are characterized by strikingly different body plans (1, 2). Because the anuran tadpole is “essentially a free living embryo” (3) adapted to feed, respirate, move, and avoid predation while undergoing drastic developmental changes, it provides a unique vertebrate model to investigate the evolutionary interaction between ontogenesis and ecology. Parallel to adult frogs, tadpoles diversified into a wide range of aquatic and semiterrestrial habitats, and now, they constitute a wide diversity of ecomorphs worldwide (4). Insights in the morphological diversification of the larval body plan are compromised by the paucity of the anuran fossil record and the long-term absence of a robust consensus for anuran phylogeny. The former is particularly true for tadpoles, because their generally small sizes and incompletely ossified skeletons reduce the chance of fossilization (1, 5). The latter was enhanced by the fact that evolutionary studies of larval morphological characters made use of conflicting phylogenetic hypotheses, some of which were inferred from the very same larval characters (6–10). Recent molecular studies using different data sources and methods have converged on very similar phylogenetic hypotheses for Anura (11–18). Together, they provide a consensus tree that allows us to explore patterns of morphological variation among extant tadpoles with unprecedented accuracy and detail.
One evolutionary scenario that seems plausible from an ecological perspective is that the morphological diversity of tadpoles increased mostly during periods of adaptive radiation (19). Recent molecular studies have shown that the rise of the anuran order was episodic and that the radiation of several lineages coincided with periods of global environmental change and biotic turnover (13, 15, 20). The radiation of four neobatrachian lineages in the Late Cretaceous/Early Tertiary, for example, marked the origin of today's largest and most diverse neobatrachian clades, which in recent taxonomic revisions (14, 21), are defined as Nobleobatrachia [also known as Hyloidea in previous studies (11, 13, 16), including (among others) Bufonidae, Dendrobatidae, Hylidae, and Leptodactylidae], Afrobatrachia (containing Arthroleptidae, Hemisotidae, and Hyperoliidae), Microhyloidea [Microhylidae sensu (13–16, 22)], and Natatanura [Ranidae sensu (13–16, 22)]. If these lineages indeed took opportunistic advantage of ecological niche availability during periods of ecological change, we would expect intensified morphological evolution corresponding with instances of increased lineage diversification.
There is an alternative indication, however, that larval morphological diversity has been largely determined by an early differentiation into basic trophic niches. In the 1950s, Orton (23, 24) defined four basic free-living tadpole morphotypes (numbered I–IV) differing mainly by oral and opercular architecture and spiracle position. Morphotype I [Xenoanura, including Pipidae and Rhinophrynidae (14)] and morphotype II (most Microhyloidea) lack keratinized mouthparts and primarily feed on suspended (planktonic) food particles. Morphotype III [Ascaphus and all known larvae of Costata, including Alytes, Bombina, and Discoglossus (14)] and morphotype IV [Anomocoela, including Megophryidae, Pelodytidae, Pelobatidae, and Scaphiopodidae (14), and most Neobatrachia except most Microhyloidea] have keratinized mouthparts adapted to scrape food particles from substrate. The four morphotypes arguably present an oversimplified picture of the extant tadpole diversity (8, 25), but the fact that three of them (I, III, and IV) characterize early-diverged lineages may indicate a basal period of intense morphological innovation.
Here, we reconstruct the history of anuran tadpole morphospace by mapping evolutionary changes in a comprehensive set of larval characters on a molecular scaffold tree inferred by integrating previous phylogenetic information with multigene analyses. To investigate observed morphospace patterns in light of the phylogenetic diversification of frogs, we compare evolutionary rates and levels of homoplasy across successive and parallel anuran radiations. Our analyses illustrate how distinct anuran radiation events differentially affected tadpole diversity and reveal a long-term pattern that cannot be explained in terms of cladogenesis alone.
Results
Evolution of Anuran Larval Morphospace.
We reanalyzed the dataset by Haas (26), which includes 131 discrete characters sampled across the major tissue types and body parts of tadpoles. Sampled taxa represent the major anuran lineages and cover a broad ecological diversity (26). The evolution of larval characters was reconstructed by constraining maximum parsimony (DELTRAN and ACCTRAN optimization) and Bayesian analyses with a molecular scaffold tree (Methods, SI Methods, and Fig. S1). Because the results were very similar for any reconstruction method, we only report on the DELTRAN-inferred patterns. The molecular scaffold tree is composed of clades that received high support by maximum likelihood (ML) bootstrapping and Bayesian analyses of 5 mitochondrial and 12 nuclear genes (Dataset S1) and clades that were consistently recovered by previous molecular studies (Figs. S2 and S3). Bayesian relaxed clock analyses of the multigene dataset are congruent with previous studies (13, 15, 16, 27) and support anuran radiation episodes in the Triassic/Early Jurassic (basal anuran radiation), Late Jurassic/Early Cretaceous (basal neobatrachian radiation), and Late Cretaceous/Early Tertiary (Afrobatrachia, Microhyloidea, Natatanura, and Nobleobatrachia) (Fig. 1A, Fig. S4, and Table S1). The younger age estimates for early anuran divergences produced by two recent studies (17, 20) are unlikely to alter the implications of our morphological analyses.
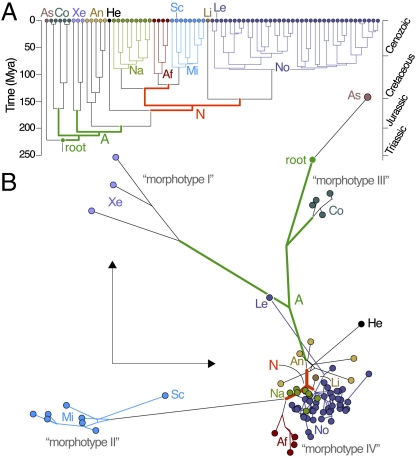
Fig. 1.
Reconstructing the evolution of tadpole morphospace. (A) Phylogenetic timescale of anuran evolution used to infer temporal patterns of tadpole evolution. Colored branches highlight the radiation episodes as defined for further analyses. A more detailed phylogeny with taxon names and clade support values is provided in Fig. S3; divergence time estimates and 95% credibility intervals are provided in Fig. S4 and Table S1. (B) 2D representation of larval morphospace obtained by multidimensional scaling. Colored circles represent positions of extant tadpoles. Branch coloration corresponds to that used in A. Internal node positions represent DELTRAN-reconstructed ancestors. A, basal anuran radiation; Af, Afrobatrachia; An, Anomocoela; As, Ascaphus; Co, Costata; He, Heleophryne; Le, Lepidobatrachus; Li, Limnodynastes; Mi, Microhyloidea; N, basal neobatrachian radiation; Na, Natatanura; No, Nobleobatrachia; Sc, Scaphiophryne; Xe, Xenoanura.
To visualize the evolutionary expansion of larval morphospace, we applied multidimensional scaling (MDS) on a pair-wise distance matrix inferred from character states of extant taxa and DELTRAN-reconstructed ancestral character states (Fig. 1B). Inspection of raw stress values (Fig. S5A) indicates that 2D was sufficient to capture the major features in tadpole morphospace without loss of essential information on pair-wise disparity. The use of 3D did not drastically alter the observed patterns (Fig. S5B).
Visual examination of the morphospace reconstruction indicates that extant tadpoles occupy a number of distinct domains, each encompassing one or several major anuran clades. Most of these domains were invaded during the basal anuran radiation in the Triassic and Early Jurassic (green branches in Fig. 1B) and locally explored during later radiation episodes. A notable exception is the domain invaded by the stem lineage of Microhyloidea, which represented a major additional morphospace extension in the Late Cretaceous. These domains are largely congruent with the taxonomic distribution of the four tadpole morphotypes described by Orton (8, 23–26). Additionally, several anuran clades, whose tadpoles are classified as Orton's morphotype IV (Anomocoela, Nobleobatrachia, Natatanura, and Afrobatrachia), occupy contiguous or even overlapping regions in morphospace. Together, these clades are estimated to contain over 80% of modern anuran species (22), implying that the majority of extant tadpoles are concentrated in a relatively small part of morphospace.
Patterns of Evolutionary Rate.
The multidirectional morphospace expansion during the basal anuran radiation followed by more localized morphospace exploration during subsequent radiation episodes suggests a deceleration of morphological diversification. Such a pattern does not necessarily imply a decline in the rate of morphological change (28, 29), because clades that show limited disparity may still have undergone high rates of morphological evolution (30). To examine how tadpole morphospace was affected by changes in evolutionary rate, we plotted the number of character changes reconstructed for successive branches through the anuran tree against their durations in millions of years (Fig. 2 A–D). Branch rates were highest during the basal anuran radiation (indicated by steep branches in Fig. 2 A–D) and fluctuated considerably along subsequent branches. Low rates characterize the neobatrachian, nobleobatrachian, and natatanuran stem branches, and a high rate characterizes the microhyloid stem branch. In addition, elevated rates of larval evolution are observed for basal branches in the neobatrachian radiation and the nobleobatrachian and microhyloid radiations. Comparison of branch rates with those inferred from simulated datasets confirms that many of the elevated branch rates depart from a null model of gradual character change across the anuran tree. Consequently, they are unlikely to represent artifacts related to stochastic rate variation, branch length patterns, or taxon sampling. Instead, these rate accelerations imply that successive episodes of increased lineage diversification left a historical footprint on the morphological evolution of tadpoles.
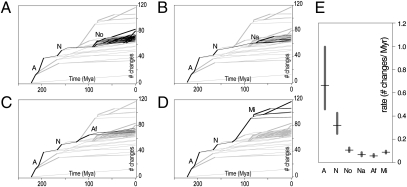
Fig. 2.
Rates of morphological evolution in tadpoles across anuran radiations. (A–D) Numbers of character changes for successive branches plotted from the root of the anuran tree to the present. The slope of each branch provides an estimate of its average rate of morphological change. For clarity, the same tree is depicted in different plots, each highlighting different root to present paths along the tree (black branches). Branches whose observed rates are significantly higher (P < 0.05) than expected under a null model of constant change are indicated in bold. (E) Rates of morphological change measured as the number of reconstructed character changes summed over all branches divided by the total branch duration. Error bars represent 95% credibility intervals incorporating uncertainty in branch duration. Abbreviations are the same as in Fig. 1.
To further compare evolutionary rates across radiations, we estimated the rate of morphological change for each defined radiation episode as the number of reconstructed character changes summed over its branches divided by its total branch duration (in millions of years). Despite the short-term rate accelerations mentioned above, our analyses show a profound difference in evolutionary rate between the basal anuran radiation and all subsequent neobatrachian radiations (Fig. 2E). Even under DELTRAN parsimony optimization (favoring most recent character changes in case of ambiguity), the average evolutionary rate of the basal anuran radiation is more than twice that of the basal neobatrachian radiation and approximately 6–12 times higher than the rates for the Late Cretaceous/Early Tertiary radiations. In summary, episodes of increased phylogenetic diversification seem to have been marked by elevated rates of morphological change but to a diminishing extent to the present.
Patterns of Homoplasy.
An additional factor that affects morphospace evolution is homoplasy, the tendency of characters to attain recurrent states by reversal, convergent, or parallel evolution. Even under frequent character changes, a lineage will fail to explore fundamentally new regions of morphospace if these changes represent reversals to ancestral conditions or parallellisms with other lineages (31, 32). Although homoplastic changes may increase tadpole diversity by generating new combinations of character states, the dimensionality of morphospace (and thus, the level of disparity among taxa) will remain limited without the exploration of new states.
We examined homoplasy across anuran radiations using the traditional homoplasy index (HI) derived from the consistency index (33) as well as a state-specific homoplasy index (HIS). For each reconstructed character state change, HIS indicates the number of times that it independently occurred elsewhere in the tree. Both indices are likely to be correlated, but HIS has the advantage over HI that it allows distinction between rare and highly recurrent state changes within a single character. Comparison of HI and HIS ranges across radiations (Fig. 3 A and B) indicates that larval evolution during the basal neobatrachian radiation and in Nobleobatrachia, Natatanura, and Afrobatrachia showed higher levels of homoplasy than in Microhyloidea and during the basal anuran radiation. A possible bias on the homoplasy scores for the recent radiations may stem from the fact that the morphological dataset was originally assembled to reconstruct anuran phylogenetic relationships (26), and thus, it may represent an undersampling of phylogenetically uninformative autapomorphies (unique state origins on terminal branches). However, reestimation of HI and HIS for these radiations after elimination of terminal branches (i.e., considering only state changes on internal branches) gave similar results (Fig. 3 A and B).
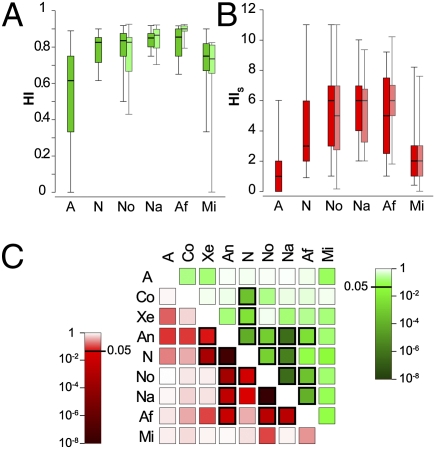
Fig. 3.
Homoplasy in tadpole morphology across anuran radiations. (A and B) Box plots represent the distribution of HI and HIS values, with boxes encompassing percentiles 25–75 and thin bars encompassing percentiles 5–95. Lighter box plots for the most recent neobatrachian radiations represent distributions when terminal branches are not taken into account. (C) Homoplasy (red boxes) and overlap in evolving characters (green boxes) shared between pairs of radiations/clades. Box coloration indicates the probability that the counted number of shared homoplastic states/evolving characters would be observed if all states/characters would evolve under equal probabilities; probabilities < 0.05 (darkest boxes) are indicated by thick frames. Abbreviations are the same as in Fig. 1.
To investigate in more detail how homoplasy is distributed across the anuran tree, we quantified homoplastic states shared pair-wise among radiations and clades. For several pairs of clades/radiations, the observed number of homoplastic changes largely exceeds the number expected if all possible character states would have had an equal probability of origination (P < 0.05) (Fig. 3C, red boxes). The most significant levels of pair-wise homoplasy are observed for Anomocoela, the basal neobatrachian radiation, Nobleobatrachia, Natatanura, and Afrobatrachia. In contrast, Microhyloidea and the basal anuran radiation share little pair-wise homoplasy with other radiations/clades, indicating that the character states that evolved along their branches were more unique with respect to the entire history of anuran evolution.
Low numbers of homosplastic states shared between the basal anuran radiation and its daughter lineages may indicate that characters that evolved new states during the basal anuran radiation may have simply reversed along secondary radiations or acquired additional states. Alternatively, however, there may have been a temporal change in the set of evolving characters when some characters that evolved early on during tadpole evolution remained invariable any time after (and vice versa). To evaluate whether low levels of shared homoplasy indeed reflect shifts in character evolution, we quantified evolving characters shared between radiations/clades, regardless of whether they produced homoplastic states (Fig. 3C, green boxes). The results strongly mirror the patterns of pair-wise homoplasy: characters that evolved during the same neobatrachian radiations and Anomocoela are broadly overlapping, but there is only limited overlap in character evolution between the basal anuran radiation and Microhyloidea and other clades/radiations. Consequently, patterns of low pair-wise homoplasy mostly reflect shifts in evolving characters rather than reversals or alternative state changes within the same set of characters.
Beyond Orton's Morphotypes.
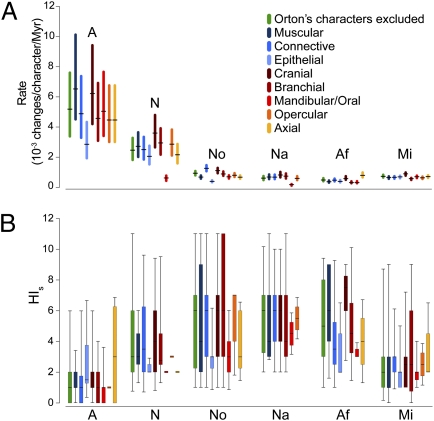
Previous evolutionary studies that referred to Orton's tadpole scheme reconstructed morphotype transitions along early branches in the anuran tree and along the microhyloid stem branch (8, 10). The presently observed morphospace extensions and shifting sets of evolving characters indeed reflect instances of major morphological change along the relevant branches. However, our analyses confirm that patterns of tadpole evolution are more complex and far-reaching than simple transitions of Orton's morphotypes. First, the observed rate and homoplasy patterns are not restricted to the characters used to define Orton's morphotypes (6–8, 10, 23–25, 34). Analyses excluding these characters yielded proportionally similar rates and homoplasy measures for all radiations (Fig. 4, green bars), with a high rate and low level of homoplasy characterizing the basal anuran radiation and low rates and high levels of homoplasy characterizing all subsequent neobatrachian radiations. Moreover, in most cases, similar results were obtained when only characters of a single major tissue type or body part were considered (epithelial and axial character sets being potential exceptions). These results suggest that the observed patterns of rate and homoplasy transcend Orton's morphotypes and rather, affect most parts of the tadpole's morphology.
Fig. 4.
Evolutionary rates (A) and homoplasy (B) in different tadpole tissue types and body parts. Error bars in A represent 95% credibility intervals incorporating uncertainty in branch duration. Box plots in B represent the distribution of HIS values of all reconstructed character changes, with boxes encompassing percentiles 25–75 and thin bars encompassing percentiles 5–95. Abbreviations are the same as in Fig. 1.
Second, the tadpoles of Lepidobatrachus and Scaphiophryne do not fit within any of the four morphotypes, which is evidenced by their distinct positions in the reconstructed morphospace (Fig. 1B). The proximity of Lepidobatrachus to the morphotype I and III domains is caused by the apparent reversal of various cranial and branchial features lost earlier in the evolution of morphotype IV tadpoles (26, 35) and the evolution of traits that are superficially similar to those of xenoanuran tadpoles (including a wide slit-like mouth, paired spiracles, and reduction of keratinized mouthparts). Unlike the morphotype II tadpoles of other microhyloids, the Scaphiophryne tadpole develops keratinized mouthparts and mandibular musculature adapted to substrate feeding, although these traits seem partially reduced compared with morphotype IV larvae (26). As a result, the Scaphiophryne tadpole has been described as a transitional form between morphotypes IV and II (26, 34). However, similar to a previous study (36), our molecular analyses support a nested position of Scaphiophryne within Microhyloidea, as a close relative of Paradoxophyla (another madagascan genus with morphotype II larvae). A Shimodaira–Hasegawa likelihood ratio test (37) favors rejection of a basal split between Scaphiophryne and the remaining Microhyloidea (P = 0.0007) but does not allow rejection of a sister clade relationship between a clade combining Scaphiophryne and Paradoxophyla, and the remaining Microhyloidea (P = 0.2101). Consequently, parsimonious explanations for the presence of mouthparts adapted to substrate feeding in Scaphiophryne imply that they were lost at least two times independently in the microhyloid radiation or that they reappeared within Scaphiophryne after loss along the microhyloid stem.
Discussion
Theoretical and empirical studies have resulted in the emergence of two recurrent themes describing the origin of morphological diversity in clades. One theme entails that morphological change and speciation are correlated (29, 30, 38). At the level of individual speciation events, this relationship forms the core of the theory of punctuated equilibria (39), and when extended to entire clades (38), it meets one of the key hallmarks of adaptive radiation (29, 40–42). A second theme arose from the observation that many clades underwent an early rapid increase in disparity followed by a slowdown of morphological diversification (43–45). These themes are often indistinguishable, because in many taxa, both morphological evolution and lineage diversification occurred disproportionately early in their evolutionary history (29, 44). However, the relative importance of both themes may become apparent in clades that underwent multiple successive episodes of lineage radiation, such as Anura.
Our results suggest that the tadpole morphospace evolved according to a mixture of the two themes: successive anuran radiations did leave a footprint on the tempo and mode of tadpole diversification, but their short-term effects are overshadowed by a major slowdown of morphological evolution shortly after the initial anuran radiation. Such slowdown in other taxa has been perceived as the result of growing niche competition and saturation of the available ecological space, increased stabilizing selection on functionally integrated traits, or establishment of intrinsic (genetic or developmental) constraints (43–47). The Late Cretaceous/Early Tertiary interval is known to be a period of global biotic turnover, and previous studies have suggested that amphibians took opportunistic advantage of contemporary ecosystem remodeling (15, 48). We find it unlikely that, during such times of ecological opportunity, diversification of tadpoles was hampered by the saturation of ecological space. In addition, many of the character changes during the basal radiation reflect adaptive differentiation into universal ecological/trophic niches (originally benthic/nektonic substrate feeding in morphotype III and IV tadpoles and midwater suspension feeding in morphotype I tadpoles). The generality of these niches seems timeless, and there is no indication that aquatic habitats imposed radically different selective pressures on the tadpole body plan during successive radiation periods.
Stabilizing selection on sets of functionally integrated characters can overcome short-term selective pressures on individual characters (49). Many of the characters used in this study play a role in three pump mechanisms that simultaneously accommodate two competing functional demands: feeding and respiration (8). In many tadpoles, the concerted action of the buccal, pharyngeal, and branchial pumps regulates both food uptake and gill irrigation, and this involves the interaction of a large number of muscles, ligaments, cartilages, and epithelial structures (50). Consequently, functionally integrated characters that were innovative during the primary radiation of a clade may remain conserved over subsequent radiations when their mutual adaptive value transcends the variation in selective pressures represented by an ecological landscape, such that their individual loss would be maladaptive in any niche. At a higher phylogenetic level, the generality of this observation seems straightforward; numerous innovations that arose during the basal metazoan radiation (e.g., triploblastic embryogenesis, central nervous system, and vertebrate skeleton) remained constant during the subsequent radiations of phyla and classes. Our analyses show that this pattern scales down to smaller clades, in which a single body plans seems evolutionarily conserved. A remaining question is whether similar patterns would characterize the evolution of free-living larvae during episodic radiations of other animal clades undergoing a distinct metamorphosis, like holometabolous insects (51).
Free-living larvae escape the physical constraints imposed by an egg during a major part of their development. This entails an increased plasticity in larval body size compared with direct developing vertebrates, both phylogenetically (e.g., the evolution of giant tadpoles in the paradoxical frog genus Pseudis) and intraspecifically [by the partial independence of larval growth and development (52)]. However, larval morphogenetic processes may still be exposed to vertical constraints caused by selective pressures on traits in adult frogs (49). In addition, more so than in direct developing vertebrates, ontogenetic processes in free-living larvae are directly exposed to ecological selection, and they are constrained to maintain a viable ecomorph throughout the entire larval period. Such tadpole-specific constraints may explain the observation that different anuran taxa show a striking level of heterochronic plasticity during early embryogenesis (53) but eventually, converge to very similar larvae when they start feeding after hatching (54).
Our study identifies homoplasy as a key descriptor of morphospace evolution in anuran larvae. In combination with a high evolutionary rate, a low degree of homoplasy along the basal anuran radiation implies the evolution of a large set of characters, many of which underwent unique or rare state changes. Conversely, the low evolutionary rates combined with high levels of homoplasy in subsequent radiations indicate small sets of evolving characters, only a fraction of which were rare or unique. As a result, homoplasy reinforced the effect of decreased evolutionary rates on morphospace evolution by simultaneously limiting its dimensionality and promoting clade overlap in morphospace. Currently, only few lineages (e.g., Microhyloidea and Lepidobatrachus) seem to have broken this pattern, although we anticipate that the analysis of additional larvae might reveal other exceptions [e.g., direct developing taxa, torrential larvae of Nasikabatrachidae (55), or megophryid larvae that retain supernumerary caudal vertebrae until shortly before metamorphosis (56)].
Microhyloidea stand out among neobatrachian radiations by showing higher evolutionary rates and lower levels of homoplasy shared with other clades. This difference suggests that the environmental or intrinsic factors that limited larval evolution in other anuran radiations had less effect on the microhyloid radiation. One explanation is that their shift to suspension feeding allowed them to escape competition with substrate feeding larvae or relaxed stabilizing selection on integrated feeding/breathing mechanisms. The parallel loss of substrate feeding also promoted homoplasy among distantly related lineages, which is not only manifested by the recurrent loss of oral structures but also by parallel heterochronic shifts among developmental processes. Larvae of Ceratophryidae (including Lepidobatrachus) and Xenoanura represent different feeding guilds (macrophagous carnivores and midwater suspension feeders, respectively), but several ontogenetic and metamorphic processes are accelerated independently in both lineages, resulting in larval conditions that anticipate postmetamorphic morphology (peramorphosis) (35, 57). A phylogenetic framework allows us to recognize homoplasy in traits and ontogenetic processes, but the integration of genomic and developmental analyses will be required to examine whether the same genetic pathways underlie their homoplastic origins (58). Similarly, insights into the genetic programs that control the development of keratinized mouthparts and related musculature may reveal whether these structures were recurrently lost during the microhyloid radiation or reversed within Scaphiophryne.
Despite lower rates of morphological evolution and higher levels of homoplasy, neobatrachian tadpoles of other radiations do not seem to have been hampered in their ecological diversification. Many neobatrachian tadpoles are generalists, and as stated by Sokol (7), “a tadpole, like that of Rana, can feed on a suspension of protists one moment, on soft or decaying macrophytes the next and on a dead horse the third.” Because of this trophic versatility, many tadpoles may not require additional morphological specialization. In addition, tadpoles that did specialize to life in particular habitats mainly did so by changes in shape, size, and number of preexisting structures (e.g., body shape, oral disk shape and size, numbers and organization of keratodonts, and tail fin height) rather than by morphological innovation. Additional specialization often involved changes in behavior, microhabitat, or developmental mode (e.g., parental care, phytothelmy, school formation, and endotrophy). Again, however, most of these specializations are rife with homoplasy across parallel radiations (19, 59).
Methods
An overview of all methods is presented in Fig. S1. Detailed method descriptions are provided in SI Methods.
Molecular Scaffold Tree.
To obtain a scaffold tree that overlapped maximally in taxon sampling with the morphological dataset of Haas (26), we used the four-step approach described in SI Methods. Briefly, we combined DNA sequences of confirmed closely related species (Fig. S2) into single chimeric taxa, each representing a corresponding taxon in the morphological dataset. The resulting matrix includes 78 chimeric anurans and sequences of 17 genes. Anuran clade support was assessed by ML bootstrapping (SBS) using RAxML 7.0.4 (60) and Bayesian posterior probabilities (BPP) using MrBayes 3.1.2 (61). Both methods implemented mixed general time-reversible (GTR +G+I) models partitioned over gene fragments. Construction of the scaffold tree was based on highly supported nodes identified by these analyses (SBS > 75% or BPP > 0.95) and consistently well-supported nodes recovered in previous phylogenetic studies (Fig. S3). Branches demarcating the basal anuran and basal neobatrachian radiations were arbitrarily defined. Alternative hypotheses regarding the phylogenetic position of Scaphiophryne were evaluated with the Shimodaira–Hasegawa likelihood ratio test (37) implemented in PAUP* 4.0b10 (62).
Morphological Analyses.
The molecular scaffold tree was used as a backbone constraint in heuristic maximum parsimony (MP) searches and Bayesian Markov chain Monte Carlo runs executed with PAUP* and MrBayes, respectively. Resulting trees were used to infer ancestral character states for the morphospace reconstruction, morphological branch lengths for the inference of rate patterns, and state changes per branch for the inference of homoplasy patterns.
Divergence Times and Branch Durations.
Divergence times were estimated using two different Bayesian relaxed clock methods implemented in MultiDivtime (63) and BEAST 1.4.8 (64). Selection of priors and time constraints is described in detail in SI Methods. Branch durations for subsequent rate analyses were calculated by subtracting for each branch the median divergence time of its terminal node from that of its parental node. Total branch durations for each radiation were obtained by summing these durations over all its branches, and corresponding 2.5 and 97.5 percentiles were calculated by repeating this for 10,000 posteriorly sampled sets of divergence times.
Morphospace Reconstruction.
DELTRAN-reconstructed ancestral states were added to the dataset of extant tadpoles to compute a pair-wise distance matrix corrected for missing data with PAUP*. MDS analyses on this matrix were performed with Statistica 8 (Statsoft). The minimal number of dimensions required to capture the essential aspects of morphospace was determined by inspecting a Scree plot and searching the nth dimension, whose addition, compared with n − 1 dimensions, caused the largest descent in raw stress (Fig. S5).
Evolutionary Rates.
Morphological branch lengths were plotted against corresponding branch durations in a cumulative way from past to present. Plots inferred from ACCTRAN and Bayesian branch lengths were similar to the DELTRAN plot (Fig. S6). To evaluate whether rate patterns were affected by sampling artifacts, we compared the DELTRAN branch lengths with those estimated under a null model of gradual change across the tree. Null distributions for branch lengths were inferred from 500 artificial datasets generated by character simulation using Mesquite 2.5 (65). Evolutionary rates for each radiation were calculated by summing character changes over all its branches and dividing this sum by its total branch duration; 95% credibility intervals were then defined by dividing these sums by the 2.5 and 97.5 percentiles of the total branch durations (yielding 2.5 and 97.5 percentiles for evolutionary rates).
Homoplasy.
HI values were derived from the credibility interval (CI) values produced per tree branch using PAUP*; HIS values were calculated manually based on DELTRAN-optimized character state reconstructions in Mesquite. Pair-wise homoplasy between two radiations/clades was evaluated based on the number of shared character state origins. Because the significance of this number depends on the total numbers of changes in each, we calculated the probability that the counted number of shared state origins would be observed if character states were randomly drawn from a constant set (SI Methods). An analogous probability was calculated for the observed number of shared evolving characters.
Supplementary Material
Acknowledgments
We thank the editor and four anonymous reviewers for valuable comments on earlier versions of this manuscript. K.R. and F.B. received postdoctoral fellowships from the Fund for Scientific Research–Flanders (FWO-Vlaanderen).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100633108/-/DCSupplemental.
References
- 1.McDiarmid RW, Altig R. Tadpoles: The Biology of Anuran Larvae. Chicago: University of Chicago Press; 1999. [Google Scholar]
- 2.Handrigan GR, Wassersug RJ. The anuran Bauplan: A review of the adaptive, developmental, and genetic underpinnings of frog and tadpole morphology. Biol Rev Camb Philos Soc. 2007;82:1–25. doi: 10.1111/j.1469-185X.2006.00001.x. [DOI] [PubMed] [Google Scholar]
- 3.Altig R. Tadpoles evolved and frogs are the default. Herpetologica. 2006;62:1–10. [Google Scholar]
- 4.Altig R, Johnston GF. Guilds of anuran larvae: Relationships among developmental modes, morphologies, and habitat. Herpetol Monogr. 1989;3:81–109. [Google Scholar]
- 5.Chipman AD, Tchernov E. Ancient ontogenies: Larval development of the Lower Cretaceous anuran Shomronella jordanica (Amphibia: Pipoidea) Evol Dev. 2002;4:86–95. doi: 10.1046/j.1525-142x.2002.01064.x. [DOI] [PubMed] [Google Scholar]
- 6.Starrett PH. In: Evolutionary patterns in larval morphology. Evolutionary Biology of the Anurans: Contemporary Research on Major Problems. Vial JL, editor. Columbia, MO: University of Missouri Press; 1973. pp. 251–297. [Google Scholar]
- 7.Sokol OM. The phylogeny of anuran larvae: A new look. Copeia. 1975;1975:1–23. [Google Scholar]
- 8.Cannatella DC. In: Architecture: Cranial and axial musculoskeleton. Tadpoles, the Biology of Anuran Larvae. McDiarmid RW, Altig R, editors. Chicago: University of Chicago Press; 1999. pp. 52–91. [Google Scholar]
- 9.Maglia AM, Pugener LA, Trueb L. Comparative development of frogs: Using phylogeny to understand ontogeny. Am Zool. 2001;41:538–551. [Google Scholar]
- 10.Pugener LA, Maglia AM, Trueb L. Revisiting the contribution of larval characters to an analysis of phylogenetic relationships of basal anurans. Zool J Linn Soc. 2003;139:129–155. [Google Scholar]
- 11.Hoegg S, Vences M, Brinkmann H, Meyer A. Phylogeny and comparative substitution rates of frogs inferred from sequences of three nuclear genes. Mol Biol Evol. 2004;21:1188–1200. doi: 10.1093/molbev/msh081. [DOI] [PubMed] [Google Scholar]
- 12.Roelants K, Bossuyt F. Archaeobatrachian paraphyly and pangaean diversification of crown-group frogs. Syst Biol. 2005;54:111–126. doi: 10.1080/10635150590905894. [DOI] [PubMed] [Google Scholar]
- 13.San Mauro D, Vences M, Alcobendas M, Zardoya R, Meyer A. Initial diversification of living amphibians predated the breakup of Pangaea. Am Nat. 2005;165:590–599. doi: 10.1086/429523. [DOI] [PubMed] [Google Scholar]
- 14.Frost DR, et al. The amphibian tree of life. Bull Am Mus Nat Hist. 2006;297:1–370. [Google Scholar]
- 15.Roelants K, et al. Global patterns of diversification in the history of modern amphibians. Proc Natl Acad Sci USA. 2007;104:887–892. doi: 10.1073/pnas.0608378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiens JJ. Global patterns of diversification and species richness in amphibians. Am Nat. 2007;170(Suppl 2):S86–S106. doi: 10.1086/519396. [DOI] [PubMed] [Google Scholar]
- 17.Blackburn DC, Bickford DP, Diesmos AC, Iskandar DT, Brown RM. An ancient origin for the enigmatic flat-headed frogs (Bombinatoridae: Barbourula) from the islands of Southeast Asia. PLoS One. 2010;5:e12090. doi: 10.1371/journal.pone.0012090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irisarri I, San Mauro D, Green DM, Zardoya R. The complete mitochondrial genome of the relict frog Leiopelma archeyi: Insights into the root of the frog Tree of Life. Mitochondrial DNA. 2010;21:173–182. doi: 10.3109/19401736.2010.513973. [DOI] [PubMed] [Google Scholar]
- 19.Bossuyt F, Milinkovitch MC. Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proc Natl Acad Sci USA. 2000;97:6585–6590. doi: 10.1073/pnas.97.12.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.San Mauro D. A multilocus timescale for the origin of extant amphibians. Mol Phylogenet Evol. 2010;56:554–561. doi: 10.1016/j.ympev.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 21.Bossuyt F, Roelants K. In: The Timetree of Life. Hedges SB, Kumar S, editors. Chicago: University of Chicago Press; 2010. pp. 279–294. [Google Scholar]
- 22.Anonymous (2011) AmphibiaWeb: Information on Amphibian Biology and Conservation. Available at http://amphibiaweb.org/Accessed April 4, 2011.
- 23.Orton GI. The systematics of vertebrate larvae. Syst Zool. 1953;2:63–75. [Google Scholar]
- 24.Orton GI. The bearing of larval evolution on some problems in frog classification. Syst Zool. 1957;6:79–86. [Google Scholar]
- 25.Duellman WE, Trueb L. Biology of Amphibians. 2nd Ed. New York: McGraw-Hill; 1992. [Google Scholar]
- 26.Haas A. Phylogeny of frogs as inferred from primarily larval characters. Cladistics. 2003;19:23–89. doi: 10.1111/j.1096-0031.2003.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 27.Van Bocxlaer I, Roelants K, Biju SD, Nagaraju J, Bossuyt F. Late Cretaceous vicariance in Gondwanan amphibians. PLoS One. 2006;1:e74. doi: 10.1371/journal.pone.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gavrilets S. Dynamics of clade diversification on the morphological hypercube. Proc Biol Sci. 1999;266:817–824. [Google Scholar]
- 29.Gavrilets S, Losos JB. Adaptive radiation: Contrasting theory with data. Science. 2009;323:732–737. doi: 10.1126/science.1157966. [DOI] [PubMed] [Google Scholar]
- 30.Adams DC, Berns CM, Kozak KH, Wiens JJ. Are rates of species diversification correlated with rates of morphological evolution? Proc Biol Sci. 2009;276:2729–2738. doi: 10.1098/rspb.2009.0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wake DB. Homoplasy: The result of natural selection, or evidence of design limitations? Am Nat. 1991;138:543–567. [Google Scholar]
- 32.Donoghue MJ, Ree RH. Homoplasy and developmental constraint: A model and an example from plants. Am Zool. 2000;40:759–769. [Google Scholar]
- 33.Kluge AG, Farris JS. Quantitative phyletics and the evolution of anurans. Syst Zool. 1969;18:1–32. [Google Scholar]
- 34.Wassersug R. The Pseudohemisus tadpole: A morphological link between microhylid (Orton type 2) and ranoid (Orton type 4) larvae. Herpetologica. 1984;40:138–149. [Google Scholar]
- 35.Fabrezi M. Morphological evolution of Ceratophryinae (Anura, Neobatrachia) J Zool Syst. 2006;44:153–166. [Google Scholar]
- 36.van der Meijden A, et al. Nuclear gene phylogeny of narrow-mouthed toads (Family: Microhylidae) and a discussion of competing hypotheses concerning their biogeographical origins. Mol Phylogenet Evol. 2007;44:1017–1030. doi: 10.1016/j.ympev.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Shimodaira S, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 38.Ricklefs RE. Cladogenesis and morphological diversification in passerine birds. Nature. 2004;430:338–341. doi: 10.1038/nature02700. [DOI] [PubMed] [Google Scholar]
- 39.Eldredge N, Gould SJ. In: Models in Paleobiology. Schopf TJM, editor. San Francisco: Freeman; 1972. pp. 82–115. [Google Scholar]
- 40.Schluter D. The Ecology of Adaptive Radiation. Oxford: Oxford University Press; 2000. [Google Scholar]
- 41.Losos JB, Miles DB. Testing the hypothesis that a clade has adaptively radiated: Iguanid lizard clades as a case study. Am Nat. 2002;160:147–157. doi: 10.1086/341557. [DOI] [PubMed] [Google Scholar]
- 42.Funk DJ, Nosil P, Etges WJ. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc Natl Acad Sci USA. 2006;103:3209–3213. doi: 10.1073/pnas.0508653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foote M. The evolution of morphological diversity. Annu Rev Ecol Syst. 1997;28:129–152. [Google Scholar]
- 44.Harmon LJ, Schulte JA., 2nd Larson A, Losos JB. Tempo and mode of evolutionary radiation in iguanian lizards. Science. 2003;301:961–964. doi: 10.1126/science.1084786. [DOI] [PubMed] [Google Scholar]
- 45.Ruta M, Wagner PJ, Coates MI. Evolutionary patterns in early tetrapods. I. Rapid initial diversification followed by decrease in rates of character change. Proc Biol Sci. 2006;273:2107–2111. doi: 10.1098/rspb.2006.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciampaglio CN. Determining the role that ecological and developmental constraints play in controlling disparity: Examples from the crinoid and blastozoan fossil record. Evol Dev. 2002;4:170–188. doi: 10.1046/j.1525-142x.2002.02001.x. [DOI] [PubMed] [Google Scholar]
- 47.Wagner PJ, Ruta M, Coates MI. Evolutionary patterns in early tetrapods. II. Differing constraints on available character space among clades. Proc Biol Sci. 2006;273:2113–2118. doi: 10.1098/rspb.2006.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vieites DR, Min M-S, Wake DB. Rapid diversification and dispersal during periods of global warming by plethodontid salamanders. Proc Natl Acad Sci USA. 2007;104:19903–19907. doi: 10.1073/pnas.0705056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson MK, Chipman AD. Developmental constraints in a comparative framework: A test case using variations in phalanx number during amniote evolution. J Exp Zoolog B Mol Dev Evol. 2003;296:8–22. doi: 10.1002/jez.b.13. [DOI] [PubMed] [Google Scholar]
- 50.Larson PM, Reilly SM. Functional morphology of feeding and gill irrigation in the anuran tadpole: Electromyography and muscle function in larval Rana catesbeiana. J Morphol. 2003;255:202–214. doi: 10.1002/jmor.10054. [DOI] [PubMed] [Google Scholar]
- 51.Wiegmann BM, et al. Episodic radiations in the fly tree of life. Proc Natl Acad Sci USA. 2011;108:5690–5695. doi: 10.1073/pnas.1012675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris RN. In: The anuran tadpole: Evolution and maintenance. Tadpoles, the Biology of Anuran Larvae. McDiarmid RW, Altig R, editors. Chicago: University of Chicago Press; 1999. pp. 279–294. [Google Scholar]
- 53.Mitgutsch C, Olsson L, Haas A. Early embryogenesis in discoglossoid frogs: A study of heterochrony at different taxonomic levels. J Zool Syst Evol Res. 2009;47:248–257. [Google Scholar]
- 54.Chipman AD. Variation, plasticity and modularity in anuran development. Zoology (Jena) 2002;105:97–104. doi: 10.1078/0944-2006-00054. [DOI] [PubMed] [Google Scholar]
- 55.Das I. Some forgotten descriptions of Nasikabatrachus (Anura–Sooglossidae) Herpetol Rev. 2007;38:291–292. [Google Scholar]
- 56.Handrigan GR, Haas A, Wassersug RJ. Bony-tailed tadpoles: The development of supernumerary caudal vertebrae in larval megophryids (Anura) Evol Dev. 2007;9:190–202. doi: 10.1111/j.1525-142X.2007.00149.x. [DOI] [PubMed] [Google Scholar]
- 57.Yeh J. The evolution of development: Two portraits of skull ossification in pipoid frogs. Evolution. 2002;56:2484–2498. doi: 10.1111/j.0014-3820.2002.tb00173.x. [DOI] [PubMed] [Google Scholar]
- 58.Wake DB, Wake MH, Specht CD. Homoplasy: From detecting pattern to determining process and mechanism of evolution. Science. 2011;331:1032–1035. doi: 10.1126/science.1188545. [DOI] [PubMed] [Google Scholar]
- 59.Altig R, McDiarmid RW. In: Diversity: Familial and generic characterizations. Tadpoles, the Biology of Anuran Larvae. McDiarmid RW, Altig R, editors. Chicago: University of Chicago Press; 1999. pp. 295–337. [Google Scholar]
- 60.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 61.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 62.Swofford DL. PAUP*Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4.0b10. Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 63.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 64.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maddison WP, Maddison DR. Mesquite: A Modular System for Evolutionary Analysis. 2008. Available at http://mesquiteproject.orgAccessed December 15, 2008.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.