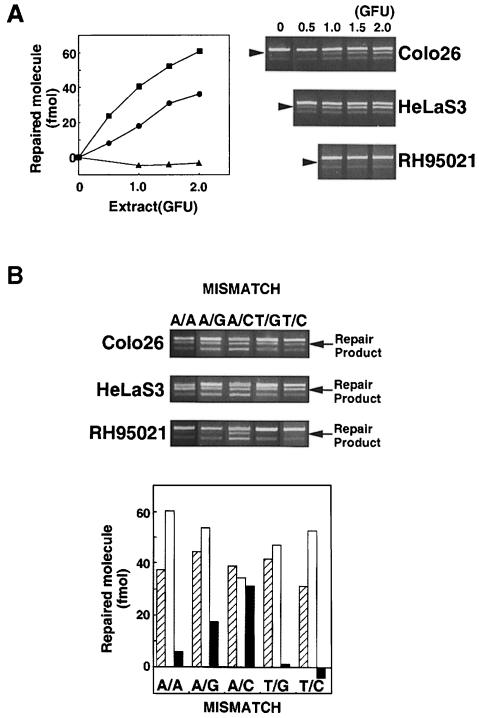
Fig. 2. Mismatch repair activity in extracts of Colo26, HeLaS3 and RH95021 cells. (A) T/C mismatch repair. Cell extracts were standardized for gap-filling activity as described in Materials and methods. The amount of extracts corresponding to the GFU indicated was incubated with the mismatched substrate (141 fmol). DNA was recovered, digested with MluI and the products separated on an agarose gel. The uppermost band is the unrepaired or incompletely repaired substrate. The repair product of 3903 bp is shown arrowed. (The lowest band represents re-annealed linearized plasmids, which do not interfere with the assay.) The amount of repair is plotted as a function of GFU for HeLaS3 (▪), Colo26 (wild-type mouse) (○) or RH95021 (Msh2–/– mouse) (▴). (B) Efficiency of correction of different mismatches by human and mouse cell extracts. Mismatch repair assays were carried out using the different mismatched substrates indicated and 1.5 GFU cell extract. After recovery of DNA and digestion with the appropriate diagnostic restriction enzyme, products were separated on agarose gels. The repair products (arrowed) were quantified as described in Materials and methods. Colo26 (□); HeLaS3 (□); RH95021 (▪).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.