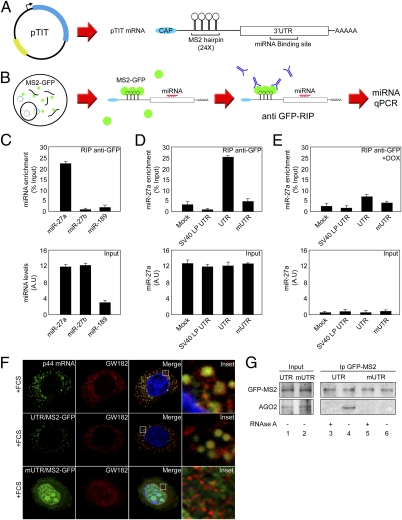
Fig. 2.
Identification of miRNAs that bind to p44 3′UTR in vivo. (A) Schematic representation of pTIT reporter system and its expressed mRNA. (B) Scheme representing miRNA identification workflow using pTIT reporter system. (C) 293-Dicer cells expressing the MS2-GFP fusion protein were transfected with pTIT-UTR, subjected to RIP with an anti-GFP antibody, and miRNA detection was assessed by RT-qPCR. Results are represented as fold-enrichment. miRNA input (27a, 27b, and 189) are depicted as control for RIP assay. (D) 293-Dicer cells expressing MS2-GFP were mock-treated or transfected with pTIT wt 3′UTR (UTR), pTIT seed region mutant 3′UTR (mUTR) or pTIT unrelated 3′UTR (SV40LP UTR) and subjected to RIP. miRNA detection was performed by RT-qPCR. Results are represented as miR-27a enrichment. miR-27a input levels are depicted as control. (E) 293-Dicer cells expressing MS2-GFP were treated with DOX, transfected as in D, and subjected to RIP. miRNA detection was performed by RT-qPCR. Results are represented as miR-27a enrichment. miR-27a input levels are depicted. (F) HeLA cells were processed for RNA-FISH against p44 mRNA (green channel) or cotransfected with an MS2-GFP expressing plasmid with pTIT-UTR or pTIT-mUTR. MS2-GFP labels pTIT produced mRNAs (green channel) and p-bodies; localization is analyzed by GW182 staining (red channel). Merged and enlarged (Inset) images are depicted. (G) 293-Dicer cells expressing the MS2-GFP were transfected with pTIT-UTR or pTIT-mUTR and used for immunoprecipitation with anti-GFP antibody. Precipitates were immunoblotted with anti-GFP (Upper) to reveal the MS2-GFP fusion protein and with anti-AGO2 (Lower). Lysates were subjected or not to RNAse A treatment (± RNase A) to analyze the RNA dependency of the interaction.