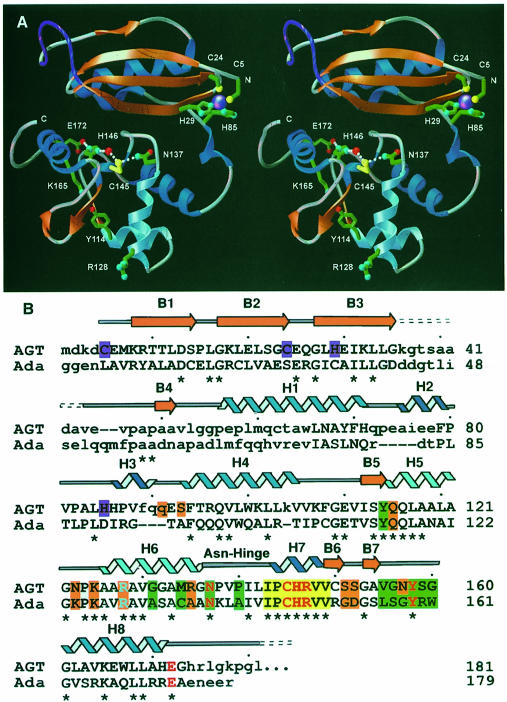
Fig. 1. Human AGT secondary structure, two-domain fold, zinc site, and location of structurally and catalytically critical residues. (A) Overall fold, displaying the active site cysteine (yellow) and its surrounding hydrogen-bond network, zinc (purple) ligands and Arg128 ‘arginine finger’. A central interdomain cleft separates the N-terminal α/β roll (α–helices in royal blue, β-strands in orange) from the C-terminal domain, which contains the active site and HTH DNA-binding motif (sky blue). For continuity, the location of the internal disordered loop (purple) has been interpolated. (B) Primary sequence, secondary structure and residue function of human AGT aligned with the E.coli Ada-C protein. Identity between the two sequences is shown with an asterisk, while corresponding residues that lie within 4.5 Å in the overlaid structures are given in upper case letters. The conserved active site motif (yellow boxes) is flanked by residues defining the O6–alkylguanine-binding channel (green boxes), anticipated DNA-binding residues (orange boxes), and HTH motif, with its associated Arg128 (sky blue). Colors distinguish residues participating in the active site hydrogen-bond network (red), zinc ligands (purple), α–helices (royal blue) and 310–helices (navy blue).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.