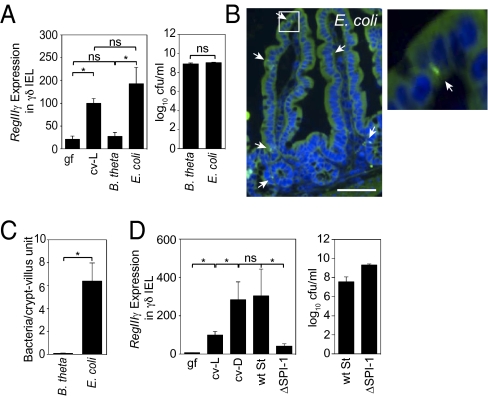
Fig. 2.
RegIIIγ expression is induced in γδ IEL by a select subset of the microbiota. (A) γδ IEL expression of RegIIIγ is triggered by a resident E. coli strain but not by B. thetaiotaomicron (B. theta). Germfree wild-type mice were gavaged with 108 cfu of B. theta or an E. coli strain isolated from the microbiota of specified pathogen-free (SPF) mice (n = 4–5 mice; representative of two experiments). After 48 h, γδ IEL were isolated and RegIIIγ mRNA was quantified by Q-PCR. Colonizing bacteria were quantified by dilution plating. ns, not significant. (B) E. coli enters small-intestinal epithelial cells. Bacteria were detected by FISH using a probe against the 16S rRNA gene (green). Arrows indicate examples of tissue-associated bacteria. Cell nuclei were visualized with DAPI (blue). As previously reported, there is nonspecific diffuse autofluorescence of intestinal tissues when visualized with FITC filters (24). (Scale bar, 50 μm.) Boxed area is enlarged at right. (C) Tissue-associated bacteria were enumerated in 200 well-oriented crypt-villus units from 5 mice per group. (D) Wild-type (wt) S. typhimurium or the invasion-deficient mutant ΔSPI-1 (108 cfu) were introduced orally into germfree C57BL/6 mice. After 48 h, γδ IEL were isolated and RegIIIγ mRNA was quantified by Q-PCR. Colonization levels were determined by dilution plating. n = 4–5 mice per group. Error bars represent ±SEM. *P < 0.05.