Figure 4.
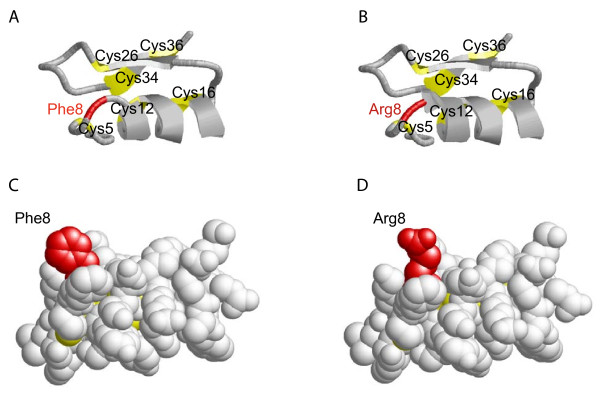
Predicted 3D structures of def1 (A) and def2 (B) peptides and the impact of amino acid substitution on molecular surface (C, D). The predicted 3D structures (A, B) show no significant differences, but the amino acids residues of Phe8 (def1) and Arg8 (def2) projected out of the molecule surface (C, D; respectively). Because the arginine carries out the positive charge, the impact on molecule def2 cationicity is without doubt. Seeing that the defensins mode of action is a thorough depolarization of the bacteria cytoplasm membrane [25], the cationicity of peptides could be the explanation for def2 higher efficiency in killing bacteria. The predicted tertiary structures of def1 and def2 peptides were based on the template 1fjnA - 3D structure of defensin MGD-1 from the mollusk Mytilus galloprovincialis[43]. Sequence similarity between MGD-1 and def1, def2 were 48.65% in both cases.