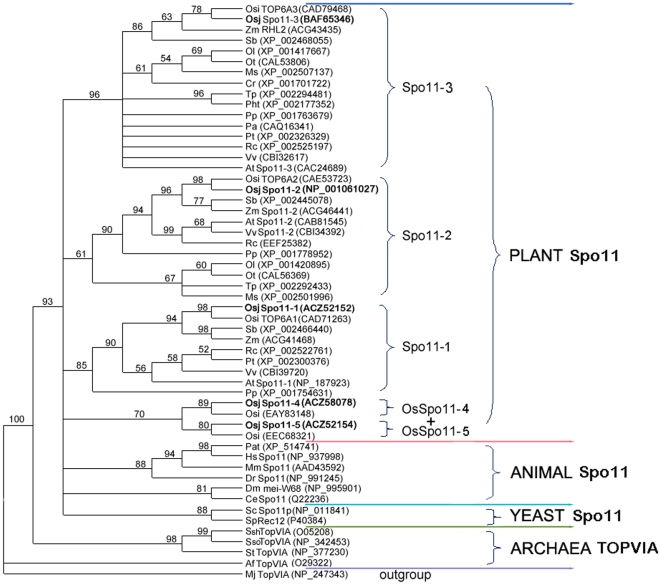
Figure 2. Unrooted Maximum Likelihood tree of Spo11/TopVIA homologues.
Sequences were obtained from the ncbi database (http://www.ncbi.nlm.nih.gov/) using the BLASTP or TBLASTN with OsSpo11-1∼5 used as quiries, and amino acids representing CAP and Toprim domain were aligned by Clustal X (version 1.8). A tree with 54 unrepeated sequences, based on a preliminary exhaustive analysis, the unambiguously aligned positions were used for tree calculation by Phyml and a JJT model of amino acid substitution. The robustness of each branch was evaluated by non-parametric bootstrap analysis for 100 replicates using Phyml software. Only bootstrap values higher than 50% are shown. MjTopVIA was selected as outgroup. Os: Oryza sativa; Zm: Zea mays; Sb: Sorghum bicolor; Ol: Ostreococcus lucimarinus; Ot: Ostreococcus tauri; Ms: Micromonas; Cr: Chlamydomonas reinhardtii; Tp: thalassiosira pseudonana; Pht: Phaeodactylum tricornutum; Pp; Physcomitrella patens; Pa: Pyrococcus abyssi; Pt: Populus trichocarpa; Rc: Ricinus communis; Vv: Vitis vinifera; At: Arabidopsis thaliana; Pat: Pan troglodytes; Hs: Homo sapiens; Mm: Mus musculus; Dr: Danio rerio; Dm: Drosophila melanogaster; Ce: Caenorhabditis elegans; Sc: Saccharomyces cerevisiae; Sp: Schizosaccharomyces pombe; Ssh: Sulfolobus shibatae; Sso: Sulfolobus solfataricus; St: Sulfolobus tokodaii; Af: Archaeoglobus fulgidus; Mj: Methanobacterium janaschii.