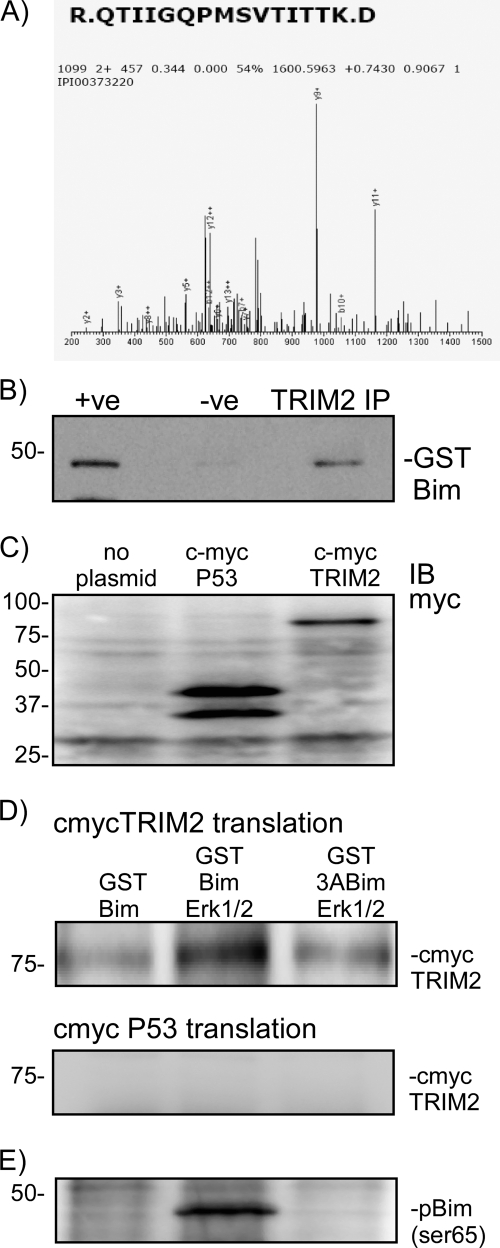
FIGURE 1.
Proteomic identification of TRIM2 as a Bim-binding protein and confirmation in a cell-free transcription/translation assay. A, phospho-Bim-binding proteins were precipitated from rat cortical neurons and subjected to one-dimensional PAGE followed by mass spectroscopy. The peptide R.QTIIGQPMSVTITK.D was identified by mass spectrometry as a phospho-Bim-binding protein. This peptide corresponds to the rat homologue of TRIM2. Because this was the only E3 ligase protein identified, it was considered further. B, binding of TRIM2 and Bim was confirmed by immunoprecipitation. Rat brain was incubated with GST-Bim and then subjected to TRIM2 immunoprecipitation. GST-Bim was identified by immunoblotting precipitated (IP) proteins with a GST-specific antibody. As a control the TRIM2 antibody was omitted from the reaction (−ve). C, using a cell-free expression system (Promega), TRIM2 and a control protein (p53 binding domain) DNA clones were synthesized into proteins. Expression was confirmed by immunoblotting (IB) lysate samples with a c-myc-specific antibody. D, GST-Bim was incubated in the absence or presence of MAPK with either myc-TRIM2 or myc-P53. As a control, a nonphosphorylatable mutant Bim (3ABim) was incubated with myc-TRIM2. Specific binding was determined by performing a GST pulldown and immunoblotting precipitated proteins for myc. Binding of myc-tagged TRIM2 increases when activated MAPK is added to the reaction. Incubation with a nonphosphorylatable BIM (3ABim) reduces this interaction. E, phosphorylation of Bim by MAPK was confirmed by blotting lysates with a Ser(P)69-specific antibody.