FIGURE 3.
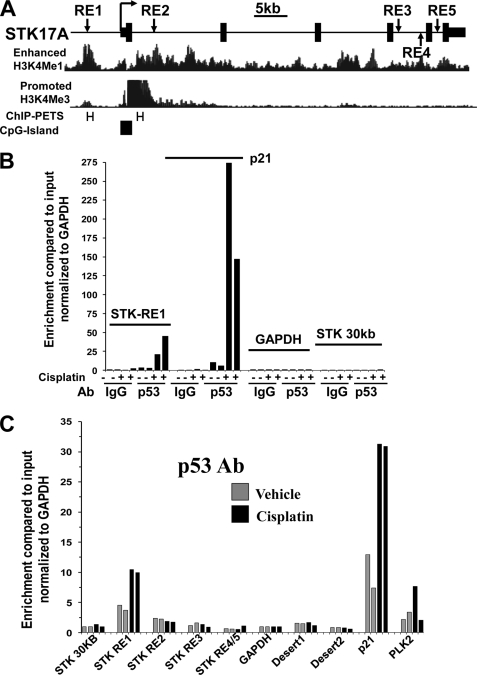
p53 binds in vivo to endogenous p53RE in STK17A. A, genomic organization of the human STK17A gene. Depicted are the locations of predicted p53-responsive elements (RE1–RE5) in relation to the transcriptional start site (arrow), exons (solid boxes), and introns (solid line) of STK17A. Positional enrichment of H3K4Me1 and H3K4Me3 modifications, p53 ChIP-PETs (PET regions are labeled with H), and CpG islands were downloaded from the UCSC Genome Browser assembly March 2006 (NCBI36/hg18). B, ChIP analysis in biological duplicate of NT2/D1 cells treated with 2.0 μm cisplatin for 6 h and harvested 10 h later. A p53 antibody, but not IgG, enriched in a cisplatin-dependent manner DNA fragments containing STK17A-p53RE1 and the well characterized p53 binding site of p21 but not for fragments of the GAPDH promoter or a region of the STK17A gene 30 kb upstream of the transcriptional start site. Real time PCR amplifications were performed for each precipitation with primers surrounding each site normalized to the signal from input DNA. C, an independent ChIP experiment under identical conditions as B with primer sets to STK17A-p53RE1 to STK17A-p53RE5 demonstrates that p53 only binds efficiently to STK17A-p53RE1 and that this was comparable with p53 binding to the previously characterized p53RE in the PLK2 gene. Note that the same primer set was used to detect STK17A-p53RE4 and STK17A-p53RE5 due to their close proximity. Two additional ChIP experiments were performed in biological duplicate and demonstrated greater than 20-fold-enrichment of p53 binding to STK17A-p53RE1.