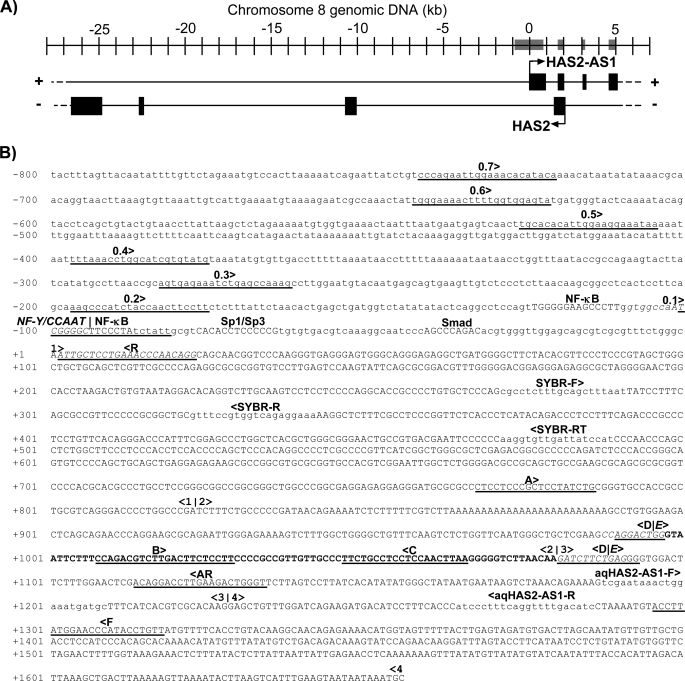
FIGURE 1.
Genomic organization of HAS2-AS1 and HAS2 at locus 8q24.13 and locations of TFBS consensus motifs and primer-binding sites for luciferase reporter, end-point RT-PCR, and qRT-PCR analyses of the human HAS2-AS1 gene. A, the four exons of HAS2-AS1 on the upper (+) strand of chromosome 8 at locus 8q24.13 are shown as black boxes, with the transcription start site indicated by an arrow. HAS2 on the lower (−) strand is depicted similarly, and the overlap of HAS2-AS1 exon 2 and HAS2 exon 1 is evident. The sequences displayed in B are highlighted in A by four gray boxes on the scale bar. B, the complete HAS2-AS1 cDNA sequence NR_002835 (26) is shown from the 5′ terminus at nucleotide +1 to the 3′ terminus at nucleotide +1656 in uppercase letters, with the following exceptions. Exon boundaries are numbered and delineated by vertical lines and arrows, e.g. <2|3> for the boundary between HAS2-AS1 exons 2 and 3. Oligonucleotide primer sequences and primer-binding site positions are displayed with orientation and labeled as described below. Primer positions for SYBR Green qRT-PCR analyses are shown in lowercase letters and are indicated as SYBR-F for the qRT-PCR sense-strand primer, SYBR-R for the qRT-PCR antisense-strand primer-binding site, SYBR-RT for the RT antisense-strand primer-binding site, aqHAS2-AS1-F for the absolute quantification qRT-PCR sense-strand primer, and aqHAS2-AS1-R for the absolute quantification qRT-PCR antisense-strand primer-binding site. End-point RT-PCR primer positions are underlined and designated A–F (including AR), with the exception of primer E, which overlaps for some of its length with primer D, and the relevant sequence is italicized. The alternatively spliced nucleotides in HAS2-AS1 exon 2 are shown in boldface type. Upstream of NR_002835 is genomic sequence from nucleotides −1 to −800. Selected discrete putative TFBSs identified by in silico analysis (30, 31) are shown in uppercase letters. The NF-Y/CCAAT site around nucleotide −100 is shown in italic type, and the overlapping NF-κB motif is shown wholly in uppercase letters. Sense-strand PCR primer sequences for the amplification of luciferase reporter construct inserts spanning ∼0.1–0.7 kb of putative HAS2-AS1 promoter sequence are underlined and designated accordingly, and the common antisense-strand primer-binding site is designated R.