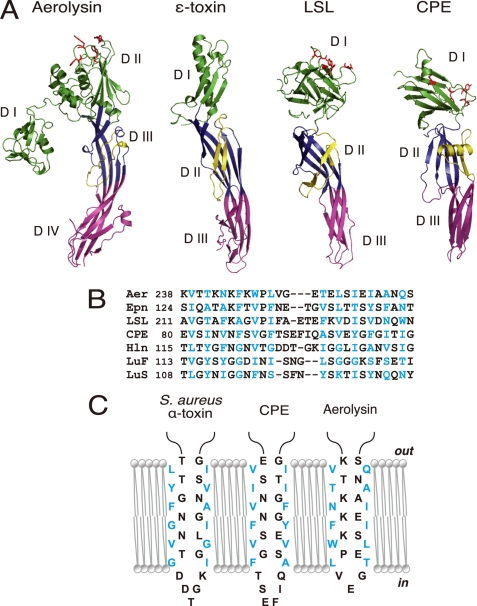
FIGURE 3.
Structural similarities between aerolysin, C. perfringens ϵ-toxin, LSL, and CPE. A, ribbon diagrams drawn from PDB data under codes 1PRE for aerolysin (17), 1UYJ for C. perfringens ϵ-toxin (35), 1W3F for LSL (36), and 3AM2 for CPE (this study). Domains corresponding to domains I, II, and III of CPE are defined by green, blue, and pink, respectively. Domains I and II of aerolysin are shown in green. Amphipathic loops (flanking elements) are indicated as yellow ribbons. The residues that participate in receptor binding are depicted as red sticks (11–13, 15, 36, 40). Note that each toxin possesses at least two long antiparallel β-strands spanning domains II and III (domains III and IV for aerolysin). B, sequence alignment of the putative transmembrane domains of different β-PFTs; the aerolysin-like family, aerolysin (Aer), ϵ-toxin (Epn), and LSL, three staphylococcal pore-forming toxins, Staphylococcal aureus α-toxin (Hln, PDB code 7AHL), LukF (PDB code 1PVL), and LukS (PDB code 1T5R), and CPE. The numbers after the toxin names indicate the N-terminal amino acid positions. Alignments were generated based on the alternating pattern of polar and hydrophobic residues. Residues that are putatively facing the lipid bilayer are shown in blue. C, schematic representation of the antiparallel strands forming the β-barrel of staphylococcal α-toxin and the corresponding residues of CPE and aerolysin. The alignment is based on previous reports (21, 37, 41) and sequence similarity. The residues are portrayed as either facing the lipid bilayer or lining the lumen of the pore.