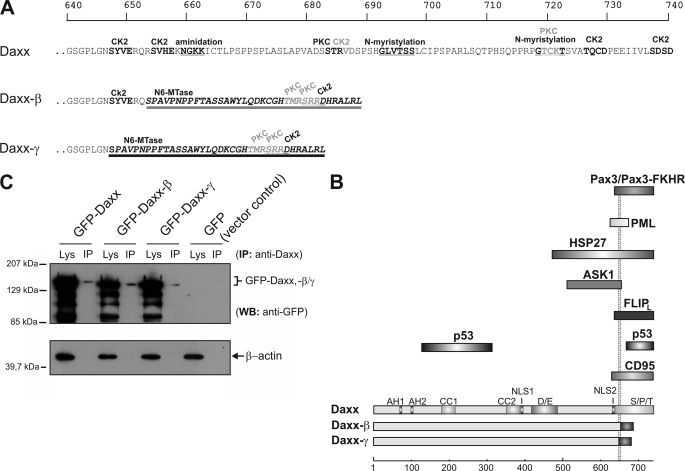
FIGURE 4.
Protein alterations of the novel Daxx isoforms. A, because of a frameshift as a consequence of the splicing event, C-terminal amino acids of the new splice variants are different from those of Daxx from amino acid residues at positions 647 (Daxx-γ, dark underlining) and 653 (Daxx-β, light underlining), respectively. Protein sequences were analyzed by PROSITE scan (34), and potential domains and modification sites are marked. Top scale resembles amino acid progression. B, schematic represents structural characteristics of the three Daxx isoforms. The respective domains are depicted in different shades of gray. Different C termini of Daxx-β/-γ are marked by dark shaded boxes. A subset of Daxx-interacting proteins together with the respective binding domain of Daxx is shown, and the two dotted lines indicate the different C termini of Daxx-β/-γ. Bottom scale represents amino acid progression. Abbreviations: CK2, casein kinase II phosphorylation site; PKC, protein kinase C phosphorylation site; N6-Mtase, N6-methyltransferase signature; Lys, crude cell extract lysate; AH, paired amphipathic helices; CC, coiled-coil regions; D/E, acid-rich domain; NLS, nuclear localization signal. C, recombinant expression of GFP-fused Daxx isoforms in HepG2 cells. GFP immunoprecipitation from vector control was performed and ran at 28 kDa (data not shown). WB, Western blotting.