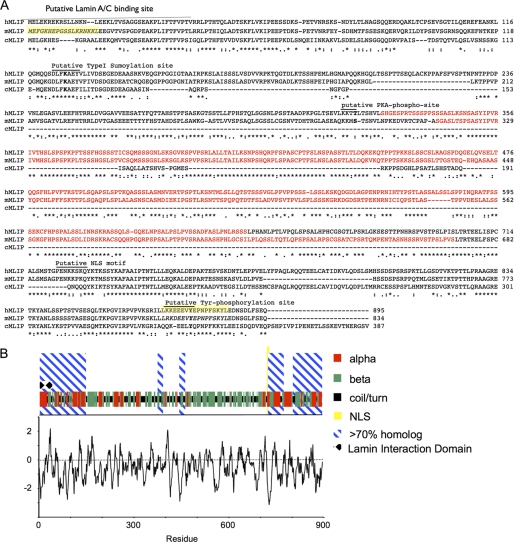
FIGURE 3.
Alignment and predicted secondary structure for human MLIP. A, the deduced amino acid sequence of human MLIP and other selected species (mouse and chicken) were aligned by ClustalW algorithm. MLIP-specific polyclonal antibodies were raised against the two highlighted (yellow) mouse MLIP peptides. The highlighted red sequence is primarily β-strand. The consensus sequence is denoted by asterisks (*) as identical, periods as conserved substitutions, and colons as semiconservative substitutions. Numbers indicate the position of the last amino acid of each line. B, the predicted secondary structure for human MLIP is represented by red, α-helical; green, β-strand; black, coil and turn. Hydrophobicity (by Kyte-Doolittle plot) of human MLIP is represented below the secondary amino acid structure of human MLIP.