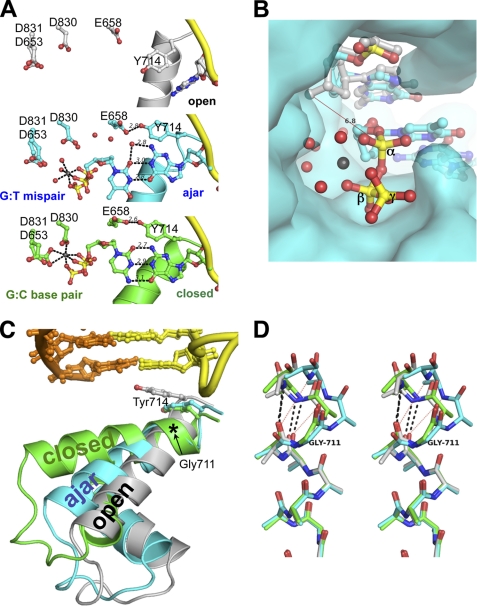
FIGURE 2.
Structures of a dG:ddTTP mismatch in the ajar Bacillus fragment active site. A, shown is a comparison between the binary BFO·DNA complex (gray; top panel), the dG:ddTTP mismatch (cyan; center panel), and the complementary dG:ddCTP base pair (gold; bottom panel). Red spheres, water molecules; gray spheres, Mg2+. B, the triphosphate of the mismatched nucleotide is misaligned for attack by the primer 3′-OH for catalysis. The open binary complex was superimposed onto the BF·DNA(dG)·ddTTP-Mg2+ crystal structure. A red line connects the modeled 3′-hydroxyl of the primer strand in the open conformation with the α-phosphate of the mismatched ddTTP. The interatomic distance is 6.8 Å. The BF polymerase is shown as its solvent-accessible surface. Red spheres, water molecules; gray sphere, Mg2+; yellow, α, β, and γ phosphorus atoms. C, the N and O helices of the fingers subdomain of BF polymerase in the open (gray), ajar (cyan), and closed (green) conformations is shown in ribbons. Tyr-714 and the post-insertion site base pair are shown in sticks. The location of Gly-711 is denoted by an asterisk. Incoming dNTPs from the ajar and closed structures have been omitted for clarity. D, structural superposition of the residues 700–710 of the O helix (in stereo) shows breakage of backbone hydrogen bonds starting at Gly-711 in the ajar conformation. Only backbone and Cβ atoms are shown. Hydrogen bonds between the backbone carbonyl oxygens of Asn-709 and Phe-710 and nitrogens of Val-713 and Tyr-714 are shown in black dashed lines for the open (gray) and closed (green) conformations. Red dotted lines show the broken hydrogen bonds in the ajar (cyan) conformation.