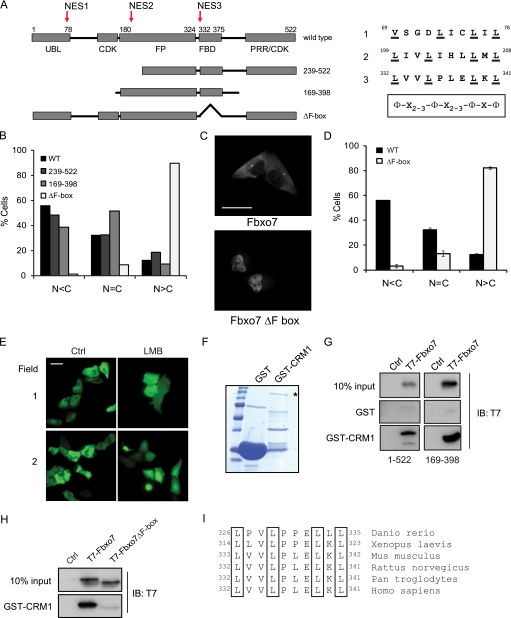
FIGURE 2.
Fbxo7 contains a functional NES within its FBD. A, schematic representation showing functional domains and truncation mutants of Fbxo7. Arrows indicate motifs within Fbxo7 that conform to the consensus sequence for a leucine-rich NES, and corresponding NES sequences are listed on the right. B, graphs of the percentage of cells showing predominantly cytoplasmic (N < C), equivalent (N = C), or predominantly nuclear (N > C) distribution of exogenous Fbxo7 proteins. Localization of Flag-tagged Fbxo7 truncation mutants was determined by immunofluorescent staining with anti-Flag antibodies. C, images of WT and Fbxo7 (ΔF-box) Venus fusions expressed in U2OS cells visualized by fluorescence microscopy. D, quantification of the distribution of the localization of WT and Fbxo7 (ΔF-box) Venus fusions from a minimum of 415 cells per condition. Error is represented as the S.E. E, fluorescence images of U2OS cells expressing a Venus-NES-Venus reporter fusion protein. Cells imaged in the right-hand side panels were treated with 20 ng/ml LMB for 15 h. F, image of the Coomassie-stained SDS-PAGE gel of GST and GST-CRM1 fusion proteins. Asterisk denotes the full-length fusion protein. G, images of immunoblots of in vitro binding assays for T7-tagged Fbxo7 (1–522) and (169–398) bound to GST-CRM1. H, immunoblots of in vitro binding assay for T7-tagged Fbxo7 (169–398) and the corresponding ΔF-box mutant binding to GST-CRM1. I, alignment of the NES amino acids located within the FBDs from Fbxo7 homologues in other species. Scale bars represent 50 μm.