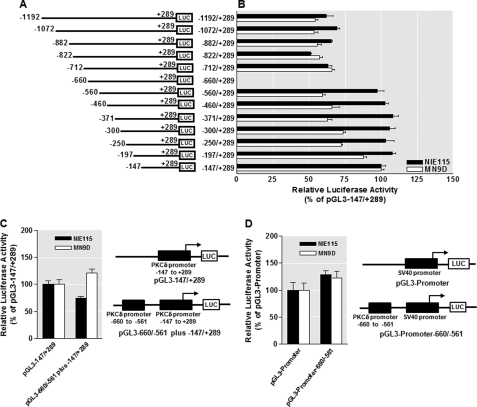
FIGURE 2.
Mapping of the identified repressive and anti-repressive elements within the PKCδ promoter and evidence for the PKCδ promoter-specific repressive element. A, schematic representation of PKCδ promoter 5′-deletion constructs used for the fine mapping study. The 5′- and 3′-positions of the constructs with respect to the transcription start site are depicted. B, each construct as depicted in A was transiently transfected into NIE115 (black bar) and MN9D (white bar) cells. Cells were harvested 24 h after transfection for assaying luciferase activities. The plasmid pcDNA3.1-βgal was cotransfected into cells for data normalization. The activity of pGL3−147/+289 was arbitrarily set to 100, and the relative luciferase activity of the other constructs is presented. The results represent the mean ± S.E. of three independent experiments performed in triplicate. C, isolated repressive element of the PKCδ promoter does not function as a locus-independent DNA element. The sequences around the identified repressive element (−660 to −561 of the PKCδ promoter) were directly fused to the 5′-end of the region between −147 to +289 of the PKCδ promoter and cloned into the pGL3-Basic luciferase vector to obtain pGL3−660/−561 plus −147/+289. NIE115 (black bar) and MN9D cells (white bar) were transfected with pGL3−147/+289 or pGL3−660/−561 plus −147/+289 for 24 h, and luciferase activity was determined. Schematic diagram of these constructs are shown at the right. The activity of pGL3−147/+289 was set to 100, and the relative luciferase activity of pGL3−660/−561 plus −147/+289 is presented. The results represent the mean ± S.E. of three independent experiments performed in triplicate. D, isolated repressive element of the PKCδ promoter does not act on a heterologous promoter (SV40). The sequences of the putative PKCδ repressive element (−660 to −561 of the PKCδ promoter) were cloned upstream of the SV40 promoter in pGL3−promoter vector to obtain pGL3−promoter-660/−561. NIE115 (black bar) and MN9D (white bar) cells were transfected with pGL3−promoter or pGL3−promoter-660/−561 for 24 h, and luciferase activity was determined. Schematic diagram of these constructs are shown at the right. The activity of pGL3−promoter was set to 100, and the relative luciferase activity of pGL3−promoter-660/−561 is given. The results represent the mean ± S.E. of three independent experiments performed in triplicate.